Brain Science
Larry H Bernstein, MD, FCAP, Curator
LPBI
A Protein Atlas of the Brain
http://www.biosciencetechnology.com/news/2015/11/protein-atlas-brain

http://www.biosciencetechnology.com/sites/biosciencetechnology.com/files/bt1511_mpi_proteome.jpg
What looks like an island is actually a schematic representation of a mouse brain. Researchers have now analyzed the mouse brain proteome and summarized the data in an atlas. (Image: MPI of Biochemistry/ K. Scharma )
Just as in the Middle Ages when there were still many uncharted areas on Earth, researchers today are aware that there is still a great deal to learn about cells in our microcosm. But instead of sextants and compasses, researchers nowadays use modern methods such as mass spectrometry to look into the world of protein molecules. Neuroscientists are focussed particularly on resolving brain complexity with its billions of specialized cells. To understand the brain’s functions, scientists from the Max Planck Institutes of Biochemistry in Martinsried and Experimental Medicine in Göttingen have for the first time quantified the entire set of proteins ‒ the proteome ‒ in the adult mouse brain. The information about which proteins and how many of them are found in the various cell types and regions has been summarized in a protein atlas.
The brain consists of hundreds of billions of interconnected cells which communicate with one another. Different cell types specialize in different functions. Nerve cells transmit and process stimuli from outside; distinct glial cells supply them with nutrients, regulate the flow of blood in the brain, help in isolating nerve fibres and perform tasks in the immune system.
Cells are comprised of proteins which are the functional building blocks. They act as small molecular machines and give the cell its structure. The information for synthesis of protein molecules is encoded in DNA and RNA; biomolecules which have been extensively examined in the brain. “Up to now, however, it was not known which and how many proteins are produced in the different, highly specialized cells or even how the numbers of proteins in the individual regions differ”, explains neuroscientist Mikael Simons. “To examine this, we needed modern measuring and analysis methods in order to be able to record and evaluate these enormous numbers of proteins.” Together with protein research specialists, a team headed by Matthias Mann in Martinsried, the scientists further developed the mass spectrometry technology for in-depth profiling of brain proteins in a rapid, reproducible and a quantitative fashion.
They were able to show that there are around 13,000 different proteins in the adult mouse brain. The quantity of proteins in the different cell types and brain regions, and how they differ from one another can now be found in the recently established protein atlas at http://www.mousebrainproteome.com. The protein data presented there from five different cell types and ten regions in the mouse brain constitute the most comprehensive collection to date.
This deep proteome investigation should serve as a rich resource for analyses of brain development and function. “Surprisingly, only 10 per cent of all proteins are cell type-specific”, explains Kirti Sharma, lead author of the study. “These cell-specific proteins are mostly found on the surface of the cell.” The large majority – 90 per cent of all proteins – are found in all cell types. As in a satellite view of previously uncharted landscapes, the researchers have created a protein atlas based on the most comprehensive data collection that should help in the development of new treatments for alleviating brain diseases.
Source: Max Planck Institute
New Computational Strategy Finds Brain Tumor-shrinking Molecules

These are MRI renderings of mouse brain tumors. Tumors treated with SKOG102 (lower panels) shrank by about half compared to tumors treated with a control (upper panels). (Credit: UC San Diego Health)
Patients with glioblastoma, a type of malignant brain tumor, usually survive fewer than 15 months following diagnosis. Since there are no effective treatments for the deadly disease, University of California, San Diego researchers developed a new computational strategy to search for molecules that could be developed into glioblastoma drugs. In mouse models of human glioblastoma, one molecule they found shrank the average tumor size by half. The study is published October 30 byOncotarget.
The newly discovered molecule works against glioblastoma by wedging itself in the temporary interface between two proteins whose binding is essential for the tumor’s survival and growth. This study is the first to demonstrate successful inhibition of this type of protein, known as a transcription factor.
“Most drugs target stable pockets within proteins, so when we started out, people thought it would be impossible to inhibit the transient interface between two transcription factors,” said first author Igor Tsigelny, Ph.D., research scientist at UC San Diego Moores Cancer Center, as well as the San Diego Supercomputer Center and Department of Neurosciences at UC San Diego. “But we addressed this challenge and created a new strategy for drug design — one that we expect many other researchers will immediately begin implementing in the development of drugs that target similar proteins, for the treatment of a variety of diseases.”
Transcription factors control which genes are turned “on” or “off” at any given time. For most people, transcription factors labor ceaselessly in a highly orchestrated system. In glioblastoma, one misfiring transcription factor called OLIG2 keeps cell growth and survival genes “on” when they shouldn’t be, leading to quick-growing tumors.
In order to work, transcription factors must buddy up, with two binding to each other and to DNA at same time. If any of these associations are disrupted, the transcription factor is inhibited.
In this study, Tsigelny and team aimed to disrupt the OLIG2 buddy system as a potential treatment for glioblastoma. Based on the known structure of related transcription factors, study co-author Valentina Kouznetsova, Ph.D., associate project scientist at UC San Diego, developed a computational strategy to search databases of 3D molecular structures for those small molecules that might engage the hotspot between two OLIG2 transcription factors. The team used the Molecular Operation Environment (MOE) program produced by the Chemical Computing Group in Montreal, Canada and high-performance workstations at the San Diego Supercomputer Center to run the search.
With this approach, the researchers identified a few molecules that would likely fit the OLIG2 interaction. They then tested the molecules for their ability to kill glioblastoma tumors in the Moores Cancer Center lab of the study’s senior author, Santosh Kesari, M.D., Ph.D.. The most effective of these candidate drug molecules, called SKOG102, shrank human glioblastoma tumors grown in mouse models by an average of 50 percent.
“While the initial pre-clinical findings are promising,” Kesari cautioned, “it will be several years before a potential glioblastoma therapy can be tested in humans. SKOG102 must first undergo detailed pharmacodynamic, biophysical and mechanistic studies in order to better understand its efficacy and possible toxicity.”
To this end, SKOG102 has been licensed to Curtana Pharmaceuticals, which is currently developing the inhibitor for clinical applications. Kesari is a co-founder, has an equity interest in and is chair of the scientific advisory board for Curtana Pharmaceuticals. Co-authors Rajesh Mukthavaram, Ph.D., and Wolfgang Wrasidlo, Ph.D., also own stock in Curtana Pharmaceuticals.
his research was funded, in part, by the National Institutes of Health, Voices Against Brain Cancer Foundation, Christopher and Bronwen Gleeson Family Trust and American Brain Tumor Association Drug Discovery Grant.
Musical Rhythms in the Brain
http://www.biosciencetechnology.com/news/2015/10/musical-rhythms-brain
Researchers at Max Planck Institute for Empirical Aesthetics in Frankfurt and of New York University have identified how brain rhythms are used to process music, a finding that also contributes to a better understanding of the auditory system. Furthermore, the study suggests that musical training can enhance the functional role of brain rhythms.
The paper, which appears in the journal Proceedings of the National Academy of Sciences, points to a newfound role the brain’s cortical oscillations play in the detection of musical sequences. The term “cortical oscillations” refers to the rhythmic electrical activity generated spontaneously and in response to stimuli by neural tissue in the central nervous system. The importance of brain oscillations in sensory-cognitive processes has become increasingly evident.
“We’ve isolated the rhythms in the brain that match rhythms in music,” explains Keith Doelling, lead author. “Specifically, our findings show that the presence of these rhythms enhances our perception of music and of pitch changes.”

http://www.biosciencetechnology.com/sites/biosciencetechnology.com/files/bt1510_maxplanck_music.jpg
Headbanging is most common in the rock, punk and heavy metal music genres. Scientist have now identified how rhythms inside the human brain are used to process music. (Image: Wikimedia / Małgorzata Miłaszewska)
Previous research has shown that brain rhythms very precisely synchronize with speech, enabling us to parse continuous streams of speech — in other words, how we can isolate syllables, words, and phrases from speech, which is not, when we hear it, marked by spaces or punctuation.
However, it has not been clear what role such cortical brain rhythms, or oscillations, play in processing other types of complex sounds, such as music.
To address these questions, the researchers conducted three experiments using magnetoencephalography (MEG), which allows the tiny magnetic fields generated by brain activity to be measured. The study’s subjects were asked to detect short pitch distortions in 13-second clips of classical piano music (by Bach, Beethoven, Brahms) that varied in tempo — from half a note to eight notes per second. The study’s authors divided the subjects into musicians (those with at least six years of musical training and who were currently practicing music) and non-musicians (those with two or fewer years of musical training and who were no longer involved in it).
Not surprisingly, the study found that musicians have more potent oscillatory mechanisms than non-musicians do. “What this shows is we can be trained, in effect, to make more efficient use of our auditory-detection systems,” observes study co-author David Poeppel, director of the Max Planck Institute for Empirical Aesthetics. “Musicians, through their experience, are simply better at this type of processing.”
For music that is faster than one note per second, both musicians and non-musicians showed cortical oscillations that synchronized with the note rate of the clips. The researchers therefore conclude that these oscillations were effectively employed by everyone to process the sounds they heard, although musicians’ brains synchronized more to the musical rhythms. Only musicians, however, showed oscillations that synchronized with unusually slow clips. This difference, the researchers say, may suggest that non-musicians are less able to process the music as a continuous melody rather than as individual notes. Moreover, musicians are able to detect pitch distortions much more accurately — as evidenced by corresponding cortical oscillations.
Thus, brain rhythms appear to play a role in parsing and grouping sound streams into ‘chunks’ that are then analyzed as speech or music, the scientists add.
Source: Max Planck Institute
New Three-Minute Test Detects Lewy Body Disease
http://www.biosciencetechnology.com/news/2015/10/new-three-minute-test-detects-lewy-body-disease
Lewy Body disease is the second most common type of progressive dementia, according to the Mayo Clinic, and affects approximately 1.3 million Americans.
The Lewy Body Dementia Association says the disease is widely accepted to be highly underdiagnosed and is the most frequently misdiagnosed form of dementia.
The new test, called the “Lewy Body Composite Risk Score” (LBCRS), developed by James E. Galvin, M.D., M.P.H., professor of clinical biomedical science at FAU, is a simple, one page-survey, that includes yes or no questions for a clinician to complete. The structured questions look at six non-motor features that are present in patients with LBD, but are much less common in other forms of dementia. The tool helps clinician assess whether a patient has rest tremor, postural instability, rigidity, or bradykensia, without having to grade each extremity.
Bioscience Bulletin: Potential Alzheimer’s Test, and Stress Linked to Stroke
N.J. Researchers Closing in on Alzheimer’s, Parkinson’s Tests
Researchers from the Rowan University School of Osteopathic Medicine, have developed a test that could detect Alzheimer’s before symptoms start by identifying a series of obscure antibodies. Scientists narrowed down the auto-antibodies he was looking at from a sample of nearly 10,000 to just 10.
Role Found for Critical Gene in 95% of ALS
Cynthia Fox interviewed experts about a recent Science study that offered new insight surrounding a protein called TDP-43 in relation to amyotrophic lateral sclerosis. The study found that in 95 percent of ALS cases the protein leaves its home – the nucleus of motor neuron cells – resulting in the creation of improper “cryptic” exons.
Like a bad teenager, in 95 percent of all amyotrophic lateral sclerosis (ALS) cases, a protein called TDP-43 leaves its home— the nucleus of motor neuron cells—to congregate, in suspect fashion, in the cytoplasm.
In a study published in Science this summer, the Johns Hopkins University (JHU) team of pathologist Phillip Wong, Ph.D., offered new insight into this molecular rebellion. It confirmed a function of normal TDP-43 in the nucleus: orchestrating proper RNA splicing and exon formation. It confirmed what lack of nuclear TDP-43 does: creates improper “cryptic” exons. And it identified proteins that mitigate effects of nuclear TDP-43 loss: potential drug leads.
“The recently published work in Science very clearly demonstrates that in the absence of TDP-43, RNA is misspliced, and in many cases targeted for degradation,” University of Michigan neurologist Sami Barmada, M.D., Ph.D., told Bioscience Technology. Barmada was uninvolved with the research. “A very real consequence of such dysfunctional RNA splicing and degradation is an inability to maintain cell health, ultimately resulting in neuron loss. The authors assembled an intriguing story that tells us quite a bit about how TDP-43 functions, and what happens to those functions in diseases such as ALS, and fronto-temporal dementia (FTD). If the mechanism identified in this manuscript does indeed underlie toxicity due to TDP-43 mislocalization, then it may very well contribute to neuron loss in the vast majority of ALS.”
High Stress Jobs May be Linked to Increased Stroke Risk
The final story in our round up this week takes a look at a new Neurology study, which found there may be a link between high stress jobs and an increased risk for stroke. After analyzing six studies, comprised of a total of 138,782 participants, researchers concluded that people with high stress jobs (such as waitresses and nursing aides) were 58 percent more likely to have an ischemic stroke than those in low stress jobs (such as natural scientists and architects).
New Scanner to Help Uncover Causes of Dementia
http://www.biosciencetechnology.com/news/2015/11/new-scanner-help-uncover-causes-dementia
The funding from the Medical Research Council will allow the SIGNA PET/MR scanner, made by GE Healthcare, to be installed in Central Manchester University Hospitals NHS Foundation Trust. Currently there are only two of these scanners in the UK, but following the Manchester funding and money to other university centers in the UK, this number will increase to seven, from a number of manufacturers.
The new scanner will help scientists and clinicians understand the causes and progression of dementia, and provide ways to test the effects of new treatments. Molecular changes in the brain are believed to be responsible for dementia and the scanners have the potential to link these with the brain changes that they cause – leading to new understanding and new treatments.
Professor Nigel Hooper is the University’s Director for Dementia Research. He said: “Dementia is a condition that is poorly understood and difficult to treat effectively. It’s going to become more of a problem in the coming decades, so our research response needs to pick up as well.
“This scanner and the wider network will give us that ability to understand dementia better and to develop more treatments.”
The scanner is expected to be operational from July 2016 and work is currently underway to refurbish a shelled space adjacent to the Nuclear Medicine Centre to house the scanner suite which will have three treatment rooms, a research office and a radiopharmacy room. CMFT was chosen as the ideal location for the scanner due to its central Manchester location and close proximity to the main University campus, the co-location of the suite adjacent to the clinical PET/CT scanner at CMFT and the opportunity to take research into clinical practice.
Christine Tonge, the Director of Medical Physics at Central Manchester said: “We are excited by this opportunity to contribute to this important area of research. This scanner will put Manchester at the forefront of dementia research and we look forward to collaborating not only with our colleagues from the University, but also from other hospitals in Greater Manchester and beyond.”
The scanner is being funded as part of the Dementias Platform UK – a multi-million pound public-private partnership, developed by the Medical Research Council, to accelerate progress in dementias research. DPUK’s aims are early detection, improved treatment and, ultimately, prevention of dementias. It is the world’s largest study group for use in dementias research, pulling together two million well-characterized participants from over 30 national population studies.
The Manchester scanner will be supervised by Professor Alan Jackson, who is the director of the University’s Wolfson Molecular Imaging Centre, which hosts two PET scanners and one MR scanner and produces radiotracers for use in PET scanning. He said: “Manchester now has a range of scanning facilities which mean that clinicians and scientists can produce high quality research across a range of conditions.
“With the growing urgency of developing treatments for dementia, this new equipment is vital in addressing a major growing health concern.
“Most importantly, being linked with the other four universities which are also purchasing PET-MR scanners will mean Manchester has the ability to become involved in multi-centre trials and research grants and contracts – increasing the effectiveness of the UK’s research in this area.”
Source: University of Manchester
Nature Reviews Neuroscience 13, 38-50 (January 2012) | http://dx.doi.org:/10.1038/nrn3121
RNA-binding proteins, and in particular TAR DNA-binding protein 43 (TDP43), are central to the pathogenesis of motor neuron diseases and related neurodegenerative disorders. Studies on human tissue have implicated several possible mechanisms of disease and experimental studies are now attempting to determine whether TDP43-mediated neurodegeneration results from a gain or a loss of function of the protein. In addition, the distinct possibility of pleotropic or combined effects — in which gains of toxic properties and losses of normal TDP43 functions act together — needs to be considered.
Transposable Elements in TDP-43-Mediated Neurodegenerative Disorders

Published: September 5, 2012 DOI: http://dx.doi.org:/10.1371/journal.pone.0044099
Elevated expression of specific transposable elements (TEs) has been observed in several neurodegenerative disorders. TEs also can be active during normal neurogenesis. By mining a series of deep sequencing datasets of protein-RNA interactions and of gene expression profiles, we uncovered extensive binding of TE transcripts to TDP-43, an RNA-binding protein central to amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Second, we find that association between TDP-43 and many of its TE targets is reduced in FTLD patients. Third, we discovered that a large fraction of the TEs to which TDP-43 binds become de-repressed in mouse TDP-43 disease models. We propose the hypothesis that TE mis-regulation contributes to TDP-43 related neurodegenerative diseases.
Citation: Li W, Jin Y, Prazak L, Hammell M, Dubnau J (2012) Transposable Elements in TDP-43-Mediated Neurodegenerative Disorders. PLoS ONE 7(9): e44099. doi:10.1371/journal.pone.0044099
Editor: Koichi M. Iijima, Thomas Jefferson University, United States of America
Accumulation of TAR DNA-binding protein 43 (TDP-43) containing cytoplasmic inclusions is a shared pathological hallmark in a broad spectrum of neurodegenerative disorders, including ALS, FTLD and Alzheimer’s disease [1]. Mutations in this multifunctional RNA binding protein are also known to underlie some familial and sporadic cases of ALS [1]. Despite considerable progress, the mechanisms that link TDP-43 to neurodegeneration still are unclear. We conducted a meta-analysis of TDP-43 protein:RNA target binding datasets and of mRNA expression datasets. All previous analyses of such data focused on sequence reads that uniquely map to the reference genome, thereby excluding transcripts derived from transposable elements (TEs). In contrast, we included sequences that map to multiple locations and examined reads that align to TEs. Our analyses lead to the striking hypothesis that TE over-expression may contribute to TDP-43 mediated neurodegeneration.
Transposable elements (TEs) are highly abundant mobile genetic elements that constitute a large fraction of most eukaryotic genomes. Retrotransposons, which copy themselves through an RNA intermediate, represent approximately 40% of the human genome [2], [3]. Although the majority of TE copies are nonfunctional, a subset have retained the ability to mobilize and even the immobile copies can be expressed [4]. Because of their potential to copy themselves and insert into new genomic locations as well as to generate enormous levels of expression, transposable elements present a massive endogenous reservoir of genomic instability and cellular toxicity [3]. The impacts of these parasitic genetic elements normally are stifled by potent cellular mechanisms involving small interfering RNAs that act via the RNA induced silencing complex (RISC) to inhibit transposon expression ([5] for review). Although most investigations have naturally focused on the germline, where new insertions are heritable and thus favored by transposon evolution, somatic tissues also have an active transposon silencing mechanism whose functional significance is less understood. An emerging literature has established that certain TEs are normally active in brain [6], [7], [8], [9] and elevated expression of some LINE, SINE (which are non-LTR retrotransposons) and LTR elements have been correlated with several neurodegenerative disorders [10], [11], [12], [13], [14], [15], [16]. We therefore investigated whether the RNA targets of TDP-43 include transposon-derived transcripts.
Several recent studies used deep sequencing to profile the RNA targets that co-purify with immunoprecipitated mouse, rat or human TDP-43 and also to profile gene expression changes in mouse after knockdown or over-expression of TDP-43 [17], [18], [19], [20]. In each case, however, these studies analyzed annotated protein coding sequences and excluded TE-derived transcripts and other repetitive elements due to the difficulties inherent in working with ambiguously mapped reads from short read technologies [e.g. [21]]. Despite efforts to develop new algorithms for analyzing multiple alignments of short reads [22], these algorithms have not been applied systematically for analyzing TE-derived transcripts in any neurodegenerative disease. Because each of the above mentioned TDP-43 related studies provided public access to their raw data, we were able to use this resource to search for TDP-43 targets and for transcript mis-expression when we included sequence reads that map to multiple genomic locations, the majority of which are TE derived transcripts in these datasets. Our meta-analysis supports three main conclusions. First, TDP-43 broadly targets TE-derived transcripts, including many SINE, LINE and LTR classes as well as some DNA elements. Second, the association between TDP-43 and TE-derived RNA targets is reduced in FTLD patients relative to healthy subjects, consistent with the idea that loss of TE control might be part of the disease pathology. Third, we observe broad over-expression of TE derived transcripts in each of two different mouse models with TDP-43 dysfunction. Finally there is a striking overlap between the TE transcripts identified as targets and those that are over-expressed with TDP-43 misexpression.
Results
We first re-analyzed raw data from the rat TDP-43 RNA immunoprecipitation sequencing (RIP-seq) dataset [17] and the mouse and human TDP-43 in vivo crosslinking-immunoprecipitation sequencing (CLIP-seq) datasets [18], [19]. We tested three different analysis methods to examine effects on TEs (Fig. 1A–C; Methods and Figs. S1 and Tables S1, S2, S3). Because reads could potentially map to many regions, we first used an analysis in which each location was weighted based on the number of alignments (Figs. 1A,B) see methods). This analysis method (MULTI), which included both unique and multi mapped reads, assigns an enrichment level for each element, but does not distinguish contributions of individual instances of each element. Although this method can potentially include effects from TEs that are difficult to map with short read sequence, a disadvantage is that it does not distinguish which instances of a given TE are detected. In addition, because many TE copies are present within introns of genes, the MULTI method does not distinguish whether the TE sequences are co-expressed with genes or expressed from TEs per se. To address these issues, and to test the robustness of our observations, we also tested two additional mapping methods for the rat and human datasets (Figs. 1C and S1E,F; Methods). First, we examined only the subset of reads that map uniquely to the genome (UNIQ). This method does bias the results to the fraction of TEs that have diverged enough to have unique sequences, but provides confidence that signal derives from unique chromosomal locations. As a third mapping strategy (UNIQ+SameEle), we examined the effects of including both uniquely mapped sequences and those that map to multiple locations so long as they map to the same element (weighted for their contribution to each instance as above – see Methods).
Magnitude (log2-fold) of enrichments (up) or depletions (down) are shown (A, rat; B, mouse) for significantly bound repeat elements grouped by class. MULTI method (see text) was used for A and B. (C) The majority of rat TE targets identified with MULTI also are identified (Left Panel, Rat) when analysis is restricted to reads that map uniquely (UNIQ) or when both uniquely mapped and multi-mapped reads that map to the same TE were included (UNIQ+SameEle). These conclusions also hold for TE targets whose binding is reduced in FTLD samples from human tissue relative to healthy controls (Left panel, Human). Most rat TE targets and differentially bound human TE targets identified from uniquely mapped reads are intergenic (Right panel). (D) For TDP-43, peaks (UNIQ+SameEle) over TE targets are tall and sharp with mean peak height of 158 counts/peak. In contrast, peak heights are lower for FUS (mean peak height of 17).
doi:10.1371/journal.pone.0044099.g001
With all three mapping strategies we find a dramatic enrichment of sequences that derive from each major class of TE (Figs. 1A–C; S1; Table S3). With the MULTI method, we find 271 significantly enriched or depleted (most were enriched) repeat element sub-families in the rat TDP-43-IP samples versus control (Fig. 1A), of which 245 correspond to TEs. In the mouse dataset (Fig. 1B), MULTI detects significant enrichment of 352 repeat element sub-families of which 334 correspond to TEs (Table S3). These comprise all major classes of TEs, including LINE, SINE, LTR and some DNA elements [3]. For instance, 85 out of the 122 known mouse LINE elements and 6 out of the 7 known rat LINE elements are identified as TDP-43 targets. Similarly 26 out of 41 mouse SINE elements and 36 out of 37 rat SINE elements also were detected as TDP-43 targets. One caveat to the mouse clip-seq analysis was the lack of a control IP to use in estimating background counts for this single dataset, which could potentially lead to a larger false positive rate in the detected peaks (see Methods); however, the similarity in the results obtained for this dataset as compared to the well-controlled studies for rat (Fig. 1A) and human datasets (see below) argues for the inclusion of this dataset despite its caveats.
Overall, we detect the most extensive binding to TEs with the MULTI method, and these findings are not an artifact of the way we assigned weights with the MULTI method because even with the more restricted UNIQ analysis, we identify ∼80% of the rat elements that are differentially enriched when all mappable reads are included (Figs. 1C, S1F). Moreover, among the uniquely mapped subset of TE instances that we identify as TDP-43 targets, greater than 80% map to intergenic regions rather than to elements contained within genes (Fig. 1C). When we include both unique mappers and multi mappers from the same element (UNIQ+SameEle), we detect enrichment for 95% of the TE sub-families that were identified as TDP-43 targets with the MULTI method (Figs. 1C, S1F). The concordant results from these three different mapping strategies provide confidence that identification of TE derived transcripts as TDP-43 targets is a robust effect that is detected with a variety of methods for dealing with multi-copy elements.
As a test of the biological specificity of our finding that TDP-43 selectively binds to TE derived transcripts, we applied the UNIQ mapping method to a CLIP-seq dataset for an unrelated RNA binding protein. For this purpose we chose fused in sarcoma (FUS), which like TDP-43, is an hnRNP RNA binding protein that plays diverse roles in RNA biology, including splicing [23]. FUS is a relevant control for specificity because like TDP-43, it is implicated in neurodegenerative disorders including ALS [24]. The result with FUS is in stark contrast with TDP-43 (Fig. 1D). For TDP-43, peaks (defined within a 500 bp window) that map to TEs are relatively large, with a mean peak height of 158 counts. In contrast, with FUS we only see small peaks over TEs with a height of just a few counts (mean peak height of 17; Fig. 1D for distribution). Peaks that map over RefGene annotations, on the other hand, are similarly distributed for both FUS and TDP-43 (Mean height of 32 and 68 respectively, Fig. S1H). The distributions of mean peak heights (see histogram, Fig. 1D) shows a clear separation between TDP-43 peaks and those obtained with FUS and this separation between peak heights is statistically significant (Wilcoxon rank sum p-value<2.2e−16). Thus our findings show specificity for TDP-43 and are not a byproduct of inherent biases in library construction or analysis.
Because TDP-43 has a known binding motif among its mRNA targets, we used MEME ([25]and see Methods) to identify enriched motifs among both the RefGene and repetitive targets. We identify a UGUGU pentamer motif that is equivalently enriched in uniquely mapped and repetitive targets (Fig. S1C; Methods). This motif is consistent with the binding specificity of TDP-43 that has previously been observed for uniquely mapped sequences [17], [18], [19], [20]. Thus TDP-43 binds TE derived transcripts via a similar sequence motif as identified for RefGene targets.
Because the human dataset [18] includes samples from healthy and FTLD patients (which exhibit TDP-43 positive cytoplasmic inclusions), it also provided an opportunity to identify differences in the TDP-43 targets between FTLD and healthy controls. As in rat and mouse, we observe in human samples a dramatic and significant enrichment in target sequences that derive from many classes of TEs. As with the mouse and rat data, the distribution of peak heights for TE and RefGene targets of TDP-43 are similar (Fig. S1I), indicating that the targeting of TE transcripts is as robust as it is for RefGene targets. More striking, however, is the comparison between healthy subjects and FTLD patients. When we examine the relative enrichment for each repeat element within healthy vs. FTLD samples, we detect a dramatic difference in binding to TE derived RNAs (Fig. 1E–H). Overall, the association between TDP-43 and TE transcripts is significantly reduced in FTLD patients, which leads to a relative enrichment of 38 repeat elements in healthy versus FTLD, 28 of which correspond to transcripts derived from TEs (Fig. 2 and Table S3; See Methods for statistical analyses). We see reduced binding of TDP-43 to transcripts from all major classes of TE including SINE, LINE, LTR and a few DNA elements. Here too, we observe that the majority of the TE targets whose binding to TDP-43 was reduced in FTLD are consistently identified with all three methods (Fig. 1C). Most of the TE targets that show reduced binding to TDP-43 in FTLD samples are intergenic rather than contained within genes (Fig. 1C). Example peaks are shown for one RefGene control (Fig. 1F) as well as two differentially targeted TEs (Figs. 1G,H).
(A) In the human CLIP-seq data from FTLD versus healthy control, 38 repeat elements showed significant (p-value< = 1e-5 and fold changes> = 2) differential binding. Log2 fold binding differences are shown for significantly enriched/depleted elements. (B,C,D) Peaks are shown in genome browser for one RefGene control (B) and two differentially targeted TEs (C,D) in Healthy (top) versus FTLD (bottom). (E) Enrichment for the UGUGU motif relative to its prevalence in the genome is shown across a 51-nt window surrounding binding sites (−25 nt, 25 nt). Healthy samples (Blue) show similar enrichment for the UGUGU pentamer motif among RefGene (solid) and repeat (dashed) sequences (RefGene/repeat motif enrichment ratio ≈1.3). In contrast, motif enrichment in FTLD samples (Red) is significantly reduced among repeat (dashed) annotations relative to RefGene (solid; p-value< = 0.01; RefGene/repeat motif enrichment ratio ≈2.0).
doi:10.1371/journal.pone.0044099.g002
This reduced binding in FTLD patients of TDP-43 to TE-derived transcripts also is apparent when we examine over-all enrichment for the UGUGU pentamer motif (Figs. 2E and S1) relative to the genome. In the rat and mouse samples as well as in the dataset from healthy human brain samples, we observe equivalent enrichment of UGUGU binding motifs among uniquely mapped (RefGene) versus repetitively mapped (repeat) TDP-43 targets (RefGene/repeat enrichment ratio near 1.0; Fig. S1D; see Methods). In the FTLD-TDP-43-CLIP samples, we also see enrichment for the UGUGU motif among RefGene targets that is equivalent to that seen in healthy subjects (Fig. 2E), but the level of enrichment for this UGUGU motif is significantly lower among the sequences that map to repeat elements. In the FTLD samples, the RefGene/repeat enrichment ratio is increased to 2.0 (Fig. 2E; p-value< = 0.01, p-values were assigned with 100 iterations on randomly chosen sets containing 50% of original data; see Methods). In other words, FTLD samples exhibit a selective reduction of binding to TE transcripts and also exhibit reduced UGUGU motif enrichment among the remaining repetitive sequences that still co-purify with TDP-43. This difference in motif enrichment between FTLD and control samples is only manifested among repeat annotations.
The reduced binding of TE transcripts in FTLD patients suggested that TDP-43 pathology might include a loss of TE regulation. We investigated this possibility in two ways. First, we analyzed the repetitive sequence reads from two different mRNA-seq datasets from mouse models of TDP-43 pathology.
The first mRNA-seq study that we analyzed [20] used over-expression of human TDP-43 in transgenic mice. Overexpression of this aggregation prone protein is associated with toxic TDP-43 pathological effects and is thought to act as a dominant-negative, causing reduction in the normal functions of TDP-43. The second mRNA-seq study [19] used antisense oligonucleotide-mediated depletion of TDP-43 in mouse striatum to test the effects of TDP-43 loss of function. Both studies identified transcripts that are differentially expressed or spliced in response to these TDP-43 manipulations. To ask if the above TDP-43 depletion and over-expression/dominant-negative impacted TE derived transcripts, we again analyzed sequence reads including those that map to multiple locations. We found broad elevations of TE derived transcripts in both the over-expression transgenic mouse model and in the striatal depletion of TDP-43 (Figs. 3A,B). TDP-43 over-expression was associated with elevated expression of 86 repetitive elements (Fig. 3A), whereas TDP-43 depletion results in increased expression levels of 223 repetitive element species (Fig. 3B). In both cases, most of these correspond to LINE, SINE and LTR elements. Overall, the affected TE transcripts are expressed at comparable levels to those of the differentially expressed RefGene transcripts (Fig. S1J), suggesting that these are robust effects on transcripts whose expression levels are not at the limit of detection. More importantly, when TDP-43 function is compromised, we observe a striking degree of concordance between the TE transcripts that are elevated and the ones that we identified as RNA targets of TDP-43 in normal tissue (Red in Fig. 3; See Table S3). Indeed the majority of elevated TE transcripts in both mouse mRNA-seq datasets also were detected as TDP-43 targets in the iCLIP-seq binding dataset (Fig. 3; Table S3). This remarkable concordance between the transcripts that are targeted by TDP-43 and those that are elevated in response to TDP-43 misexpression is unique to the repetitive elements in the genome. In contrast, CLIP targets identified from the RefGene fraction of the transcriptome have little overlap with those that show over-expression when TDP-43 function is compromised suggesting that the coding gene expression increases are largely indirect effects [19]. RefGene transcripts whose expression is reduced show good concordance with direct target identification.
(A,B) Over-expression [20] of TDP-43 in transgenic mice and depletion [19] of TDP-43 in mouse striatum each result in elevated expression of many TE derived transcripts. The majority of over-expressed TEs also were detected (Table S3) as binding targets by CLIP-seq (RED). A few showed elevated expression but were not detected as binding targets (BLUE).
doi:10.1371/journal.pone.0044099.g003
TDP-43 aggregation and neuropathology plays a fundamental role in a broad spectrum of neurodegenerative disorders [1], [26], [27]. This hnRNP-like RNA binding protein already has been implicated in a remarkable number of cellular functions including repression of HIV-1, alternative splicing, regulation of mRNA stability and microRNA biogenesis [26], [27]. Importantly, a large number of cellular targets of TDP-43 have been characterized, leading to the hypothesis that one key role of this multi-functional protein is to regulate alternative splicing of mRNA targets with a preference for those with large UG rich introns [17], [18], [19], [26], [28]. Our findings support the novel hypothesis that TDP-43 also targets the mobile element derived transcriptome. This association is defective in FTLD patients and the TE transcriptome is broadly over-expressed in mouse models of TDP-43 pathology.
A large fraction of the genetic material of multicellular organisms is made up of mobile elements as well as inactivated TEs. A fraction of these TEs retain the capacity to copy themselves and insert at new genomic locations. During the co-evolution of TEs with their host genomes, organisms have evolved elaborate and efficient mechanisms to prevent or at least regulate such transposition events. As a result, even the potentially active TE copies rarely mobilize within the germline and are also largely constrained in somatic tissue. Several recent studies demonstrate, however, that LINE-1 elements are normally active and mobile during neurogenesis in both rodent and human tissue [7], [8], [9]. Somatic mobilization of Alu and SVA elements as well as LINEs also has recently been detected in several different human brain regions [6]. This raises the intriguing hypothesis that active mobilization of some TEs plays a role in normal brain development or physiology. On the other hand, there also is emerging evidence that unregulated activation of TEs is associated with neuropathology. TE activation in brain has been observed in macular degeneration [14], Rett syndrome [11], Prion diseases [13],[29], Fragile-X associated tremor/ataxia syndrome (FXTAS) [15] and ALS [12]. Moreover, for the cases of macular degeneration and FXTAS, there is evidence that activation of SINEs and an LTR-retrotransposon respectively may contribute to the observed pathology [14], [15].
Our findings support three conclusions. First, that TDP-43 broadly targets TE-derived transcripts, including many SINE, LINE and LTR classes as well as some DNA elements. This conclusion is replicated in three independent datasets from rat, mouse and human. Second, the association between TDP-43 and TE-derived RNA targets is reduced in FTLD patients relative to healthy subjects, consistent with the idea that loss of TE control might be part of the disease pathology. Third, we observe broad over-expression of TE derived transcripts in each of two different mouse models with TDP-43 dysfunction. And there is a striking overlap between the TE targets identified in the CLIP study and those that are over-expressed with TDP-43 misexpression. Taken together, our findings raise the hypothesis that TDP-43 normally functions to silence or regulate TE expression. When TDP-43 protein function is compromised, TEs become over-expressed. Unregulated TE expression can have a number of detrimental impacts including genome instability, activation of DNA-damage stress response or toxic effects from accumulation of TE-derived RNAs or proteins. Such toxicity from activation of mobile genetic elements may contribute to TDP-43-mediated neurodegenerative disorders.
Does a loss of TDP-43 function cause neurodegeneration?
Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, 364 Plantation St, 817 LRB, Worcester, MA, 01605, USA
Molecular Neurodegeneration 2012, 7:27 doi:10.1186/1750-1326-7-27 http://www.molecularneurodegeneration.com/content/7/1/27
In 2006, TAR-DNA binding protein 43 kDa (TDP-43) was discovered to be in the intracellular aggregates in the degenerating cells in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), two fatal neurodegenerative diseases [1,2]. ALS causes motor neuron degeneration leading to paralysis [3,4]. FTLD causes neuronal degeneration in the frontal and temporal cortices leading to personality changes and a loss of executive function [5]. The discovery triggered a flurry of research activity that led to the discovery of TDP-43 mutations in ALS patients and the widespread presence of TDP-43 aggregates in numerous neurodegenerative diseases. A key question regarding the role of TDP-43 is whether it causes neurotoxicity by a gain of function or a loss of function. The gain-of-function hypothesis has received much attention primarily based on the striking neurodegenerative phenotypes in numerous TDP-43-overexpression models. In this review, I will draw attention to the loss-of-function hypothesis, which postulates that mutant TDP-43 causes neurodegeneration by a loss of function, and in addition, by exerting a dominant-negative effect on the wild-type TDP-43 allele. Furthermore, I will discuss how a loss of function can cause neurodegeneration in patients where TDP-43 is not mutated, review the literature in model systems to discuss how the current data support the loss-of-function mechanism and highlight some key questions for testing this hypothesis in the future.
Amyotrophic lateral sclerosis (ALS) is a disorder where progressive degeneration of large motor neurons in the spinal cord and cerebral cortex leads to paralysis and death [3,4]. Frontotemporal lobar degeneration (FTLD) causes degeneration of neurons in frontal and temporal cortices, leading to deterioration of executive, cognitive and social functions, as well as loss of emotional control[5]. Although clinically distinct, a significant overlap exists between these two diseases in the patient population, resulting in a continuous spectrum ranging from patients with one disease at either end and patients with varying degrees of both diseases in the middle [6,7]. Recent genetic data has reaffirmed the connection between these two diseases. Some genetic mutations cause one disease but rarely the other, e.g. SOD1, FUS and TDP-43 for ALS, and tau, progranulin and CHMP2B for FTLD. Other mutations cause either or both diseases in the same patient or family, e.g. ubiquilin 2 and C9ORF72. In a significant population of patients (~95 % ALS and ~50 % FTLD), TDP-43 positive intracellular inclusions are present in the CNS even though the TDP-43 gene is not mutated [8–11], raising the question of how wild-type TDP-43 is involved in the pathogenesis of these cases.
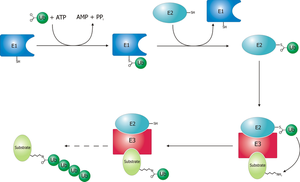
TDP-43 is a RNA binding protein containing two RNA-recognition motifs (RRM), a nuclear localization signal (NLS) and a nuclear export signal (NES) [12]. The protein is normally concentrated in the nucleus but also shuttles back and forth between the nucleus and cytoplasm[13]. TDP-43 is a global regulator of gene expression and is involved in regulation of transcription and multiple aspects of RNA processing and functioning, including splicing, stability, transport, translation and microRNA maturation [14–17]. TDP-43 interacts with many proteins and RNAs and functions in multi-protein/RNA complexes [18–21]. TDP-43 maintains its protein expression at a constant level within a tight range by auto-feedback mechanisms, which involve TDP-43 binding to its own 3’ untranslated region [15,22]. Overexpression of TDP-43 leads to down-regulation of the endogenous TDP-43 [23,24], and blocking expression of one allele leads to a compensatory increase in the expression of the other allele [25–27]. The tight regulation of TDP-43 levels is suggestive of its crucial role in the functioning of multi-protein/RNA complexes, where maintaining a certain stoichiometry between TDP-43 and the other components may be critical.
Because mutations in TDP-43 lead to ALS, a causal role of TDP-43 for neurodegeneration is firmly established [12,28,29]. Therefore, understanding how the mutants cause neurodegeneration offers a convenient entry point for exploring how TDP-43 plays this role. The first question is whether a gain, a loss of function or a dominant-negative effect mediates neurotoxicity. A resolution to this question is of critical importance because it sets the direction of further research on the disease mechanism and on the design of therapeutic strategies. To answer this question, model systems of both gain or loss of function must be employed (Table 1). Gain-of-function models are usually achieved by gene overexpression and loss-of-function models by gene knockout or knockdown. Based on the phenotypic readouts, the mechanism whereby the mutants cause neurodegeneration can be deduced (Table 1).
Table 1.Assay for disease mechanism using transgenic animals
A gain-of-function (Table 1, GF column) mechanism includes two scenarios: first, the mutant gene gains a novel toxic activity that is independent of the normal function of the gene, and second, the mutant becomes hyperactive in one of its normal functions leading to toxicity. In the first scenario, overexpression of the mutant gene, but not the wild type, will cause the disease phenotype. In the second scenario, overexpression of either the mutant or wild-type gene will cause the disease phenotype. In both gain-of-function scenarios, knockout or knockdown of the gene is not expected to cause the disease phenotype.
A loss of function (haploinsufficiency; Table 1, LF column) means that the mutant gene has no function or a reduced function but does not interfere with the function of the wild-type allele. In this scenario, neither overexpression of the mutant nor the wild type is expected to cause the disease phenotype. But knockout or knockdown reproduces the loss of function, and therefore, is expected to generate the disease phenotype.
A dominant-negative mechanism (Table 1, DN column) denotes the condition where the mutant allele is dysfunctional and inhibitory to the function of the wild-type allele. In this scenario, overexpression of the mutant gene is expected to cause the disease phenotype because it dominant-negatively inhibits the function of the endogenous wild-type protein. On the other hand, overexpression of the wild type is generally not expected to generate the disease phenotype because the wild-type gene can function normally and does not inhibit the function of the normal endogenous allele. However, there are exceptions under certain circumstances, for example, if the protein functions in a multi-protein complex (see details below). Knockout or knockdown of the gene is expected to reproduce the disease phenotype because this reduces the function of the wild-type gene. Thus, in model systems, the dominant-negative mechanism can display characteristics of both a gain and a loss of function—it is a loss of function in essence, yet its effect can dominate over the endogenous wild-type allele.
In the case of TDP-43, an abundance of gain-of-function models have been generated in various species, including worm, fly, fish and rodents [12]. In all models with rare exceptions, a consistent finding is that overexpression of both mutant and wild type TDP-43 can cause a neurodegenerative phenotype (Table 1, TDP-43 columns), thus supporting a gain-of-function mechanism and a potential overactivation of TDP-43 in the mutants [12]. Loss-of-function models have also been generated in non-mammalian species and all except the worm showed neurological and neurodegenerative phenotypes [30–33,35,44]. The difference between worm and the other species may reflect some species difference, since TDP-43 is dispensable for survival in the worm but not so in other species. In general, the degenerative phenotypes in the loss-of-function models appear less overwhelming than the overexpression models and are often difficult to separate from the developmental effects stemming from a lack of TDP-43 function. Importantly, there is a lack of evidence in mammalian models that a loss of TDP-43 function causes neurodegeneration. This is largely due to the failure in generating such a model using a gene knockout approach [25–27,36]. As a result, the current literature leans towards a gain-of-function mechanism as far as the role of TDP-43 in neurodegeneration is concerned.
Yet despite the preponderance of evidence for the gain-of-function mechanism, it has not been sufficient to rule out the loss-of-function mechanism, because the gain-of-function mechanism does not explain well a phenomenon that is consistently observed in numerous pathological studies, i.e. the nuclear clearance of TDP-43 that accompanies the presence of TDP-43 intracellular aggregates[1,2,45]. The question whether the depletion of TDP-43 in the nucleus is consequential in the pathogenesis remains unanswered. In addition, although the aggregates in the cytoplasm may generate gain-of-function type of toxicity, it is also conceivable that the aggregation of TDP-43 renders TDP-43 non-functional, and as such, causes TDP-43 dysfunction. In this review, I propose a model that is centered on the loss-of-function mechanism whereby TDP-43 plays its role in neurodegeneration. I will highlight the evidence in the current literature that is consistent with this model and the evidence that is still needed from future experiments to test this model.
A model for the loss of TDP-43 function as a central mechanism of pathogenesis in human disease
The TDP-43 protein is normally expressed through transcription and translation, and once produced, it regulates its own expression by a feedback mechanism, i.e., upregulating its own expression when the protein level is too low and inhibiting its expression when the protein level is too high [15,22–27]. By this auto-regulatory mechanism, the intracellular level of TDP-43 is maintained within a narrow range (Figure 1, #1 normal). This tightly maintained TDP-43 level may be important because TDP-43 functions in multiprotein/RNA complexes [18–21], where a proper structure and function of the complex requires a certain stoichiometric ratio between TDP-43 and its protein and RNA partners (Figure 1, #1 normal). Such a requirement is not unique to TDP-43 complexes as it has been demonstrated in other protein-RNA or protein complexes. For example, in the primary micro RNA (pri-miRNA) processing Drosha complex, overexpression of one subunit DGCR8 leads to an inhibition in the processing activity [46]. As another example, in the kinesin-2 heterotrimeric complex that drives the antegrade transport of late endosomes and lysosomes, overexpression of one subunit KAP3 inhibited the transport similar to the KAP3 knockdown [47].

Xu Molecular Neurodegeneration 2012 7:27 doi:10.1186/1750-1326-7-27 Download authors’ original image
http://www.molecularneurodegeneration.com/content/figures/1750-1326-7-27-1.gif
In the disease situation, conditions in patients’ cells become conducive for TDP-43 aggregation. For example, TDP-43 mutants and its C-terminal fragments associated with ALS and FTLD have enhanced aggregation propensity [48–51], and therefore, can drive TDP-43 aggregation. The aggregation can lead to a reduction in the pool of TDP-43 that can be incorporated into the TDP-43 protein/RNA complexes (Figure 1, #2 aggregation), thereby reducing the complex function and leading to neurodegeneration.
In model systems where TDP-43 is overexpressed (Figure 1, #3), the function of TDP-43 can be inhibited because an oversupply of exogenous TDP-43 mismatches with a limited supply of its endogenous interacting protein/RNA partners, resulting in the formation of incomplete and dysfunctional complexes. Below I highlight the evidence in the current literature that is consistent with this model and the future experiments that are need to test this model.
TDP-43 performs functions of vital importance, but the consequence of its dysfunction in neurodegeneration remains unclear
A crucial piece of evidence for a loss-of-function mechanism would be demonstration that a loss of TDP-43 function can cause neurodegeneration. This has not yet been experimentally achieved in a convincing manner, particularly in mammalian species. Knockouts in rodents cause early embryonic lethality [25–27,36]. Inducible knockout in adult mice causes a rapid loss of fat tissue and lethality [36]. These results have not been informative as to the consequences of TDP-43 dysfunction in the nervous system. Nevertheless, the severity of the phenotype in the knockout models suggests a critical functional importance of TDP-43 in the health and survival of mammalian cells. Indeed, the conditional knockout of TDP-43 in mouse embryonic stem cells causes cell death [36]. Therefore, it is conceivable that TDP-43 function may also be vital for the survival and function of neurons. Supporting this notion are the experiments where TDP-43 knockdown causes morphological abnormalities and cell death in cultured neurons [50,52,53] and a large change in gene expression in cells of the CNS [15,16].
Experimental data from non-mammalian species have also been consistent with the critical functional importance of TDP-43. In C. Elegans, TDP-43 deletion mutants are viable, but show low fertility, slow growth and locomotor defects [44]. In Drosophila, TDP-43 knockout causes abortive embryonic development and lethality [30,31]. Although some escape the lethality and develop to adults, they display severe locomotor defects, premature death and abnormal neuronal morphology [30,31]. Evidence for progressive axonal degeneration and locomotor defects has also been reported in adult TDP-43 knockdown flies [32]. In zebrafish, TDP-43 knockdown during embryonic development causes selective defects in motor axonal growth and results in motor behavioral abnormalities [35]. These results do not conclusively demonstrate a role of TDP-43 dysfunction in neurodegeneration in ALS and FTLD, but do indicate that TDP-43 is important in the development and functioning of the nervous system, thus leaving open the possibility that TDP-43 dysfunction could play a role in neurodegeneration.
How a loss of TDP-43 function explains the pathogenic mechanism of TDP-43 mutants
Mutations in TDP-43 cause motor neuron degeneration and ALS [28,29]. The overwhelming majority of the mutations are located in the C-terminal glycine-rich domain [12], which is unstructured and responsible for interactions with other proteins [17,21,54]. How mutant TDP-43 causes neurodegeneration is not known. Overexpression models support a gain of function, but the reliance of overexpression to elicit neurodegenerative phenotypes risks over-interpretation. A lack of convincing evidence that TDP-43 levels are elevated in human disease leaves open the question of whether the results from the overexpression models are relevant for the human disease.
While there is room for doubt for the gain-of-function mechanism, evidence for the loss-of-function mechanism is also weak, primarily because few experiments have generated data directly relevant to this question, especially in mammalian systems. Nevertheless, reasonable scenarios for this mechanism can be formulated based on the current, albeit fragmented and incomplete, experimental literature. First, wild-type TDP-43 is an aggregation-prone protein and mutant TDP-43 is even more so [48,51,55]. Therefore, TDP-43 mutants can initiate and drive protein aggregation, leading to TDP-43 depletion from the cell nucleus, as has been observed in patients [1,2,56]. In addition, mutant TDP-43 may have an enhanced susceptibility for polypeptide fragmentation, which generates the patient-specific 25-kDa fragments [29,57]. These fragments have a high propensity for aggregation [50,55,58] and can coaggregate with wild-type TDP-43, thereby sequestering wild-type TDP-43 into the aggregates and depleting TDP-43 from the nucleus [50].
Second, the mutant may be functionally less active or inactive but may still retain its autoregulation capability. As a result, the overall TDP-43 level would be maintained but the function of TDP-43 would be reduced because the protein expressed from the mutant allele is dysfunctional. Some experimental data support this scenario. In mice, overexpression of mutant TDP-43 inhibited the expression of the endogenous TDP-43 to the same extent as wild type overexpression [23,37,38], suggesting that the disease-causing mutants retain their autoregulatory function. In Drosophila, wild-type TDP-43 is capable of promoting growth of dendrites and increasing the size of synaptic terminals at the neuromuscular junction. However, these activities are lost in the ALS-causing mutants [31,34], suggesting that the mutants have lost some of the wild-type functions.
Third, mutant TDP-43 may form defective TDP-43 protein/RNA complexes, thereby poisoning the function of the complex. In this capacity, the mutant TDP-43 can act dominant-negatively to inhibit the function of the wild-type allele. There is evidence that TDP-43 forms a homodimer [59] and that multiple TDP-43 molecules are incorporated into each complex [19]. Therefore, if a mutant TDP-43 molecule were capable of rendering dysfunction to the whole complex that contains both mutant and wild-type TDP-43 molecules, then the function of the wild-type allele would be inhibited.
These scenarios are consistent with a model where TDP-43 mutants cause a loss of TDP-43 function by a dominant negative mechanism. Notably, while the first scenario requires the formation of aggregates for cellular toxicity, the second and third scenarios make such a requirement unnecessary. Indeed, in both cellular and animal models, toxicity induced by mutant TDP-43 does not require its aggregation [33,37,39,60].
How TDP-43 dysfunction could contribute to neurotoxicity from overexpression of either mutant or wild-type TDP-43 in model systems
The prevailing interpretation for the observation that overexpression of mutant TDP-43 causes neurodegeneration is that mutant TDP-43 exert its toxicity by a gain of function. However, these results are also consistent with a dominant-negative mechanism, as discussed above (also see Table 1). The dominant-negative model predicts that overexpression of the mutant in sufficient quantities will inhibit the function of the two endogenous wild-type alleles in the model systems.
A puzzling observation is that overexpression of wild-type TDP-43 causes similar neurotoxic phenotypes in model systems [23,33,35,37,38,40–43,60,61]. Because of the autoregulatory mechanism, overexpression of human wild-type TDP-43 leads to a suppression of the endogenous TDP-43 [23,24]. This has led to a proposal that a loss of the endogenous TDP-43 caused neurotoxicity [24]. While this proposal can reasonably explain the toxicity of the mutants on the premise that they are dysfunctional, the toxicity from the wild-type TDP-43 poses a problem because several studies have shown that the human wild-type TDP-43 gene can substitute the function of its homologue in species as distant as Drosophila and C. Elegans[30,44]. A more plausible explanation can be derived from the fact that TDP-43 functions in multiprotein/RNA complexes, whose function may depend on a certain stoichiometric composition of the different protein/RNA components. Overexpression of wild-type TDP-43 provides an amount of TDP-43 in excess of the other components that form the complexes, thereby sequestering those components into incomplete and dysfunctional complexes (Figure 1, #3 overexpression). Therefore, both overexpression of the mutants and the wild-type TDP-43 can cause neurodegeneration by dominant-negatively inhibiting the normal function of TDP-43 complexes so long as it interacts with two or more components in the complexes simultaneously and with near equal binding affinities.
While the above interpretation of the literature remains to be confirmed by further experimentation, some of the predictions from this loss-of-function/dominant-negative hypothesis are supported by observations in the current literature. First, overexpression of mutant should be more potent in causing neurodegeneration than overexpression of the wild type, which has been the case in several overexpression models [35,40,60,61]. Although this finding is not inconsistent with the gain-of-function mechanism, the result can also be explained readily by the dominant-negative mechanism outlined above. Overexpression of mutants can inhibit normal TDP-43 function by three mechanisms: (1) displacing the endogenous TDP-43 through the autoregulation mechanism, (2) inserting itself into the TDP-43 complexes in the place of the wild-type protein, and (3) forming dysfunctional complexes by disruption of the stoichiometry between TDP-43 and other protein/RNA components. In contrast, overexpression of the wild-type TDP-43 can inhibit TDP-43 function only through the third mechanism because unlike the mutant protein, it has full function. Therefore, to inhibit TDP-43 function to the same degree, a higher level of expression will be required for the wild-type TDP-43 than the mutant.
Second, if the dominant-negative hypothesis is correct, overexpression and knockout or knockdown of the gene can cause similar phenotypes. Currently, data from mammalian species is lacking to address this point. However, evidence can be drawn from other species. For example, overexpression of either mutant or the wild-type TDP-43 in Drosophila motor neurons causes progressive locomotor defects and a shortening of lifespan [33]. These phenotypes are similar to those caused by TDP-43 knockdown [33]. As another example, expression of human TDP-43 mutants but not the wild type in zebrafish embryos compromised motor axonal growth and caused locomotor defects. Similarly as in flies, knocking down the endogenous TDP-43 caused the same phenotypes [35]. Importantly, the phenotypes in the knockdown fish are rescued by the expression of human wild-type TDP-43 but not the mutants. These results are consistent with the view that the ALS-relevant TDP-43 mutants are dysfunctional and are capable of inhibiting TDP-43 function in a dominant negative manner.
Third, the loss-of-function/dominant-negative hypothesis predicts that ALS-causing mutants should be loss-of-function alleles. As discussed above, the observations that the mutants lost their ability to stimulate the growth of dendrites and axons in flies [31,34,35] and their inability to rescue phenotypes from TDP-43 knockdown in zebrafish [35] supports the loss-of-function proposition. However, key evidence from mammalian species remains to be produced.
While the case for a loss of function by a dominant-negative mechanism can be argued for, it may be overly simplistic to argue that a gain of function does not contribute to the phenotypes caused by TDP-43 overexpression in the model systems. Some evidence indicate that TDP-43 is capable of causing cellular toxicity by a gain of function under ectopic and overexpressed conditions. For example, TDP-43 causes toxicity in yeast, which does not possess an endogenous TDP-43 homologue [62]. Similarly, TDP-43 is not essential in C. Elegans, yet overexpression of human TDP-43 can still cause toxicity that is not observed in knockouts [44,61,63,64]. Therefore, in model systems where TDP-43 performs vital functions, phenotypes caused by TDP-43 overexpression are likely derived from both an interference of endogenous TDP-43 function and a gain of function. Given the complexity in the protein/RNA interaction networks of TDP-43, perhaps this would not be surprising. Overexpression is likely to generate new aberrant interactions as well as to disrupt the authentic interactions that are vital for the cell. Therefore, disentangling these effects will be complex in the overexpression models.
What is the role of wild type TDP-43 in human neurodegeneration
While the case for a loss of function in the TDP-43 mutants and in the overexpression model systems can be made, can the loss-of-function mechanism play a role in patients where TDP-43 is not mutated and not overexpressed? This is an important question because the vast majority of patients with ALS and FTLD-TDP do not have TDP-43 mutations. The answer to this question is yes because even though the primary trigger of the degenerative process lies not in TDP-43 but elsewhere, the same kind of TDP-43 aggregation and nuclear clearance is observed in the CNS of these patients [1,2,45] (Figure 1). The loss-of-function/dominant-negative model will predict that the nuclear clearance and the cytoplasmic aggregation of TDP-43 are probably a significant contributor to neurodegeneration by causing a loss of TDP-43 function. However, the experimental data for testing this prediction is scarce. In Drosophila and zebrafish, knockout or knockdown of TDP-43 produced similar neurodegenerative phenotypes [33,35]. However, further analysis is needed to differentiate the effects of TDP-43 dysfunction on neurodegeneration from those on neurodevelopment, and the relevance of these observations to human neurodegeneration remains to be established. A mammalian model with TDP-43 dysfunction in the mature CNS is urgently needed to understand the effects from a loss of TDP-43-function.
Based on the loss-of-function/dominant-negative hypothesis outlined above, what triggers TDP-43 aggregation will be one of the most intriguing and important questions in understanding the pathogenic mechanisms in ALS and FTLD. Recent investigations have shown that multiple causes can trigger secondary TDP-43 aggregation and nuclear clearance. These causes can be classified into several categories: (1) Gene mutations that enhance the mutant protein aggregation propensity and cause ALS-FTLD with TDP-43 aggregation. Examples in this category include VCP, optineurin, dynactin, ataxin 2 and ubiquilin 2. All the mutant proteins form aggregates and some form coaggregates with wild-type TDP-43 [9,65–69]. The mechanism whereby these mutants cause TDP-43 aggregation is not understood. One possibility is that the aggregation of these proteins weakens the capacity of cellular proteostasis [70], which creates an environment conducive for aggregation-prone proteins such as TDP-43 to aggregate. Some of the proteins such as VCP and ubiquilin may be involved in TDP-43 degradation [71,72]. Therefore, mutations in these proteins may directly alter the TDP-43 economy and cause TDP-43 aggregation. (2) Gene mutations that cause ALS and FTLD with TDP-43 aggregation, but the mutant proteins are not involved in protein aggregation themselves. Examples in this category include progranulin, angiogenin and C9ORF72[1,11,73,74]. At present, it is not known how these mutations lead to TDP-43 aggregation. (3) Traumatic brain injury that lead to ALS-FTLD without gene mutations. Repetitive traumatic brain injury has been shown to be associated with ALS and FTLD with intracellular TDP-43 aggregation[75,76]. (4) Other neurodegenerative diseases that are not ALS-FTLD but trigger secondary TDP-43 aggregation. Examples of this category include some of the most common neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease and numerous others [8,77,78] (Figure1). Aggregation of TDP-43 in these cases may also be attributed to a disruption of proteostasis environment due to the aggregation of other proteins, although direct experimental evidence for this hypothesis is not yet in existence. (5) Unknown causes in sporadic ALS and FTLD cases. Some of the speculated causes include genetic predisposition in combination with environmental stress, e.g. environmental toxins, trauma and high physical activity [79–82].
Recent studies have suggested that a redistribution of TDP-43 to the cytoplasm may be a precursor to TDP-43 aggregation. In ALS and FTLD patients, some neurons show an increase in cytoplasmic TDP-43 immunoreactivity with diffused or granular appearance, which may represent an early stage of TDP-43 aggregation [83–86]. The cause for the cytoplasmic redistribution is not clear. However, a recent study demonstrate that a single traumatic brain injury can be followed by a persistent increase in the cytoplasmic levels of TDP-43 [87], suggesting that injuries to the CNS can be an initial trigger for increased levels of cytoplasmic TDP-43. In model systems, the redistribution of TDP-43 can be triggered by various stresses, including neuronal injury [88–90], overexpression of disease-associated mutant TDP-43 and VCP [91–93], oxidative stress [93,94] and proteasome inhibition [53]. The functional consequence of the cytoplasmic localization of TDP-43 will require further characterization. Nevertheless, some studies suggest that the cytoplasm-localized TDP-43 is recruited to stress granules before being transformed into aggregates that can persist independent of stress granules [93–95]. Another study demonstrated that a modest knockdown of TDP-43 exacerbated, rather than alleviated, cell death that is induced by proteasome inhibition and associated with TDP-43 cytoplasmic translocation [53], suggesting that any toxicity that might be associated with TDP-43 cytoplasmic translocation is derived from a loss of TDP-43 function. These data are consistent with the hypothesis that an increased cytoplasmic level of TDP-43, which follows the initial cellular stress, can lead to TDP-43 aggregation and nuclear depletion.
Therapeutic implications from the dominant-negative model
Discussion on therapeutic implication based on the loss-of-function hypothesis may be premature since the hypothesis remains to be tested. However, such an exercise may be helpful for illustration of the critical importance for a resolution of this question. In the case of a gain of function, strategies that reduce the function should be effective. This may be achieved by lowering the protein levels through an inhibition of its synthesis or a stimulation of its degradation. If the toxic activity is known, strategies that inhibit the specific toxic activity may also be effective. In the case of a loss of function, on the other hand, strategies that increase the function should be effective. This may be achieved by increasing expression and stability of the protein, or stimulating its activity.
The therapeutic strategy for the dominant negative mechanism differs from both purely gain- or loss-of-function mechanisms and will be most challenging. We cannot simply increase the level of TDP-43 because uncontrolled increase of TDP-43 may inhibit the function of TDP-43 rather than improving it. High levels of TDP-43 could also further accelerate its aggregation and produce aberrant interactions with other proteins and RNA. Moreover, we do not understand why TDP-43 stays in the cytoplasm and becomes depleted from the nucleus in the disease. Therefore, it is not clear whether a simple increase of TDP-43 will replenish its level in the nucleus. In the case of mutant TDP-43, allele-specific inhibition of the mutant TDP-43 may be helpful but may not be sufficient to compensate for the lost function of the mutant allele. If the hypothesis that TDP-43 aggregation drives nuclear depletion of the TDP-43 is correct, preventing or reversing the aggregation may be a rational and safe approach to mitigate the loss of TDP-43 function. To achieve this, we need to understand how TDP-43 aggregation is triggered and propagated. We also need to understand the TDP-43 aggregation process at molecular and structural levels. Alternatively, strategies that enhance the function of TDP-43 without resorting to increase the protein level, or retain TDP-43 in the nucleus may also be effective.
Conclusions
TDP-43 aggregation and nuclear depletion have been observed widely in neurodegenerative diseases. The role of TDP-43 in neurodegeneration remains to be defined. Chief among the questions is whether a gain of function, a loss of function or a dominant-negative mechanism is responsible for neurotoxicity. The answer to this question is of critical importance because it guides the future direction of research and sets the foundation for therapeutic strategies. Current experimental data from model systems has been predominantly invoked to support the gain-of-function mechanism. However, a careful review of the data suggests that a loss of TDP-43 function caused by its mutations, its aggregation and nuclear depletion, and the inhibition of TDP-43 function by a dominant-negative mechanism in the overexpression models, are at least as plausible as the gain-of-function theory, if not more so. Therefore, in our future research, we need to gain a more detailed understanding of the normal function of TDP-43, particularly in the cells of the CNS. We need models of loss of TDP-43 function in the CNS, particularly in mammalian species, to understand the consequence of TDP-43 dysfunction. In such a pursuit, models with a partial loss of TDP-43 function may be especially desirable because in humans, it is unlikely that the TDP-43 function is totally lost. We need evidence from human diseases to determine whether the conditions are more in tune with a gain or a loss of TDP-43 function. Lastly, we need to design strategies to address the difficult problem of how to restore the normal levels of TDP-43 function as a therapy.














 worms, that reducing the activity of the signaling mechanism conveyed through insulin and the growth hormone IGF1, a major aging regulating pathway, constituted a defense against the aggregation of the Aβ protein which is mechanistically-linked with
worms, that reducing the activity of the signaling mechanism conveyed through insulin and the growth hormone IGF1, a major aging regulating pathway, constituted a defense against the aggregation of the Aβ protein which is mechanistically-linked with