Upregulate Tumor Suppressor Pathways
Writer and Curator: Larry H Bernstein, MD, FCAP
7.5 Upregulate Tumor Suppressor Pathways
7.5.1 NR4A nuclear receptors are orphans but not lonesome
7.5.2 The interplay of NR4A receptors and the oncogene–tumor suppressor networks in cancer
7.5.3 NLRX1 acts as tumor suppressor by regulating TNF-α induced apoptosis
7.5.4 The Mre11 Complex Suppresses Oncogene-Driven Breast Tumorigenesis and Metastasis
7.5.5 Expression of Stromal Cell-derived Factor 1 and CXCR4 Ligand Receptor System in Pancreatic Cancer
7.5.6 DLC1- a significant GAP in the cancer genome
7.5.7 DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma.
7.5.8 Smad7 regulates compensatory hepatocyte proliferation in damaged mouse liver and positively relates to better clinical outcome in human hepatocellular carcinoma
7.5.1 NR4A nuclear receptors are orphans but not lonesome
Kurakula K, Koenis DS, van Tiel CM, de Vries CJ.
Biochim Biophys Acta. 2014 Nov; 1843(11):2543-2555
http://dx.doi.org/10.1016/j.bbamcr.2014.06.010
Highlights
- Nuclear receptors Nur77, Nurr1 and NOR-1 are ‘orphan’ receptors of the NR4A subfamily.
- The NR4A receptors have no ligands.
- The known protein–protein interactions of all three NR4A receptors are summarized.
- Interacting proteins are transcription factors, coregulators or protein kinases.
- Protein–protein interactions modulate NR4A receptor activity and function.
The NR4A subfamily of nuclear receptors consists of three mammalian members: Nur77, Nurr1, and NOR-1. The NR4A receptors are involved in essential physiological processes such as adaptive and innate immune cell differentiation, metabolism and brain function. They act as transcription factors that directly modulate gene expression, but can also form trans-repressive complexes with other transcription factors. In contrast to steroid hormone nuclear receptors such as the estrogen receptor or the glucocorticoid receptor, no ligands have been described for the NR4A receptors. This lack of known ligands might be explained by the structure of the ligand-binding domain of NR4A receptors, which shows an active conformation and a ligand-binding pocket that is filled with bulky amino acid side-chains. Other mechanisms, such as transcriptional control, post-translational modifications and protein–protein interactions therefore seem to be more important in regulating the activity of the NR4A receptors. For Nur77, over 80 interacting proteins (the interactome) have been identified so far, and roughly half of these interactions has been studied in more detail. Although the NR4As show some overlap in interacting proteins, less information is available on the interactome of Nurr1 and NOR-1. Therefore, the present review will describe the current knowledge on the interactomes of all three NR4A nuclear receptors with emphasis on Nur77.
Nur77 in the regulation of endocrine signals and steroid hormone synthesis
Nur77 is expressed in endocrine tissues and in organs that are crucial for steroid hormone synthesis such as the adrenal glands, the pituitary gland and the testes. The first functional NurRE was identified in the promoter of the pro-opiomelanocortin (POMC) gene of pituitary derived AtT-20 cells [2]. Nur77 can bind this NurRE either as a homodimer or as a heterodimer with either one of the other two NR4A receptors Nurr1 and NOR-1. Interestingly, it was shown that these heterodimers enhance POMC gene transcription more potently than homodimers of Nur77 do, suggesting that there is interdependency between the NR4A receptors in activating their target genes [3]. The NurRE sequence in the POMC promoter also partially overlaps with a STAT1-3 response element. Philips et al. showed that Nur77 and STAT1-3 bind simultaneously to this so called NurRE-STAT composite site and synergistically enhance transcription of the POMC gene. However, Nur77 and STAT1-3 do not interact directly, which suggests that oneor more facilitatingfactors are involved in NurRE-STAT driven transcription. Mynard et al. showed that this third factor is cAMP response element binding protein (CREB), which binds both STAT1-3 and Nur77 and indirectly enhances transcription of the POMC gene by facilitating the synergistic activation of the NurRE-STAT composite site by STAT1-3 and Nur77 [4]. Nur77also plays animportant role in the steroidogenic acute regulatory protein (StAR)-mediated testosterone production by Leydig cells. StAR is required for the transport of cholesterol through the mitochondrial membrane to initiate steroid hormone synthesis. Nur77 binds to an NBRE in the StAR promoter, which is in close proximity to an AP-1 response element. In response to cAMP stimulation c-Jun and Nur77 synergistically increase StAR gene expression [5], presumably through a direct interaction between c-Jun and the LBD of Nur77 [6]. On the other hand, c-Jun has also been shown to suppress expression of the hydroxylase P450 c17 gene by blocking the DNA-binding activity o fNur77 in response to stimulation of Leydig cells with reactive oxygen species [7].The effect of c-Jun on the transcriptional activity of Nur77 therefore seems to depend on other factors as well. One of these factors could be the atypical nuclear receptor DAX1 (NR0B1), which lacks a DBD and associates with multiple coregulatory proteins. DAX1 binds Nur77 directly and represses its ability to enhance transcription of the previously mentioned P450 c17 gene.
Fig.1.Schematic representation of the domain structure of nuclear receptors. Nuclear receptors are composed of an N-terminal domain (N-term), a central DNA-binding domain (DBD) and a ligand-binding domain (LBD). The amino acid similarity between the individual domains of Nur77 with Nurr1 and NOR-1 is given in percentages below the domains.
http://ars.els-cdn.com/content/image/1-s2.0-S0167488914002134-fx1.jpg
The interactome of NOR-1
NOR-1 is less well studied than Nur77 and Nurr1 and most of the data on interacting proteins of NOR-1 are presented in studies that are mainly focused on its homologues. As a consequence, NOR-1 protein– protein interactions are described with limited detail, for example the HATp300/CBPacetylatesNOR-1similarlyasNur77,however,theeffect on NOR-1 activity has not been described [79]. Likewise, NOR-1 interacts with the co-regulator TIF1β resulting in enhanced NOR-1 activity, but the domain involved in the interaction is unknown [48]. Similar to Nur77, PKC and RSK1/2 were shown to induce NOR-1 mitochondrial translocation [73,79] and DNA-PK binds the DBD of NOR-1. Even though Nurr1 and Nur77 are both essential for optimal DSB repair the function of NOR-1 in this process remains to be studied [68]. Both FHL2 and the peptidyl-prolyl isomerase Pin1 bind the N-terminal domain and DBD of NOR-1, resulting in reduced or enhanced transcriptional activity of NOR-1, respectively [59,64]. Muscat and co-workers performed detailed studies to identify coregulatorsofNOR-1andwerethefirsttorevealtheabsenceofaconventional ligand-binding pocket in the LBD of NOR-1, through molecular modeling and hydrophobicity analysis of the LBD [104]. Based on these analyses, the relative importance of the N-terminal domain of NOR-1 in regulation of the transcriptional activity of NOR-1 became apparent and direct interaction of a number of crucial co-regulators to this domain was shown;SRC-2 (GRIP-1), SRC-1, SRC-3, p300, DRIP250/ TRAP220 and PCAF [104]. The interaction between the N-terminal domain of NOR-1 and TRAP220 is independent of PKA- and PKC phosphorylation sites in TRAP220. Most interestingly, the purine derivative 6-mercaptopurine, which enhances the activity of NR4As without directly binding these nuclear receptors promotes the interaction between NOR-1 and TRAP220 [105]. Both Nur77 and NOR-1 are involved in T-cell receptor mediated apoptosis of developing T cells [106]. During activation of T cells the expressionofNOR-1isinducedandproteinkinaseC(PKC)becomesactive.NOR-1is aPKCsubstratethat isphosphorylatedand subsequently translocatesfromthenucleustothemitochondriawhereitbindsBcl-2. Most interestingly, as already indicated above the interaction between NOR-1/Nur77 and Bcl-2 causes a conformational change in Bcl-2 allowing its BH3 domain to be exposed, resulting in the conversion of Bcl-2 from an anti-apoptotic into a pro-apoptotic protein. For Nur77 it is exactly known which amino acids are involved to provoke the functional switchin Bcl-2, whichis not thecasefor NOR-1 [57,79]. Initially, the homeobox domain containing protein Six3 was identified in a yeast-two-hybrid study as a protein that interacts uniquely withtheDBDandLBDofNOR-1withoutbindingorinhibitingtheactivity of Nur77 or Nurr1. Of interest, NOR-1 and Six3 show overlap in expression in the rat fetal forebrain on embryonic day 18 [107]. In a later study this specificity of Six3 forNOR-1 was not found, rather interaction with all three NR4As was observed [108]. NOR-1 is part of the EWS/NOR-1 fusion protein that is expressed in human extraskeletal myxoid chondrosarcoma tumors. Six3 enhances the activity of NOR-1 (and Nur77 and Nurr1), whereas the activity of EWS/NOR-1 is inhibited and the interaction only requires the DBD of NOR-1. The opposing data in these two studies may be explained by the use of different cell types for the activity assays, as well as the use of Gal4-fusion proteins in the latter study. PARP-1 specifically and effectively interacts with theDBD of NOR-1 independent of the enzymatic activity of PARP-1 [69]. Nurr1 interacts with lower affinity, whereas EWS/NOR-1 and Nur77 do not bind PARP-1, unless the N-terminal domain of Nur77 is deleted. The latter experiment nicely illustrates that the N-terminal domains of Nur77 and EWS/NOR-1 disturb PARP-1 interaction with the DBD. This may be the underlying mechanism of differential function of NOR-1 and the EWS/NOR-1 fusion protein. In line with the binding characteristics, PARP-1 only inhibits the activity of NOR-1 effectively, again independently of the ribose polymerase activity of PARP-1.
Table 5 NOR-1 interacting proteins.
Fig.2. Nur77 and its interacting proteins. Schematic overview of the protein–protein interactions with Nur77 for which the domains of interaction have been elucidated. Details are described in the text and in Tables 1–3, which also contain the full names of the indicated proteins. N-term, N-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain.
http://ars.els-cdn.com/content/image/1-s2.0-S0167488914002134-gr2.sml
Fig.3. Nur77 and kinases modulating its activity and localization. A, Overview of the amino-acid sequence of Nur77 with known phosphorylation sites and associated kinases indicated (T= threonine,S= serine). B,Schematic illustration of effects of different kinases on Nur77 transcriptional activity and subcellular localization. See Table3 for definitions of the abbreviations of the kinases shown.
http://ars.els-cdn.com/content/image/1-s2.0-S0167488914002134-gr3.sml
Discussion and concluding remarks
This review summarizes the currently available knowledge on the protein–protein interactions of the NR4A nuclear receptor family and their downstream effects. When looking at the information gathered in this review three main observations can be made. First, there are a large number of protein–protein interactions that regulate the activity of Nur77 and there is a large variation in the effects of these interactions on the ‘target’ protein, be it Nur77 or the interacting protein itself. These effects include modulation of transcriptional activity, protein stability, post-translational modification and cellular localization: all processes that are tightly regulated by ligand binding in other nuclear receptors. In light of the many interactions it undergoes with other proteins, Nur77 could also be considered to be a molecular ‘chameleon’: a protein that selectively adopts the responsiveness of other proteins by directly interacting with them. Secondly, the protein–protein interactions with Nur77 described in this review have been studied in a wide range of cell types, such as immune cells (T-cells, thymocytes, monocytes and macrophages); somatic cells(neurons,smooth muscle cells,endothelial
cells and hepatocytes) and cancer cells from diverse origins.We reason that a stimulus- and cell type-specific expression pattern of interacting proteins may be decisive in determining both the interactions of NR4 As with other proteins and their activity in general.The well-studied interaction between Nur77 and RXRα, which has unique outcomes depending on both the cell type studied and the stimulus used, is one such interaction that is modulated by stimulus- or cell type- specific auxiliary proteins. Lastly, there is a large amount of overlap in interacting proteins between the three NR4A nuclear receptors. All three domains of the NR4As are involved in interactions with other proteins (Tables 1–5, Fig. 2), and we think that the unstructured N-terminal domains are of special interest as they have the lowest overall amino acid similarity (Fig. 1). Based on this dissimilarity, it could be hypothesized that the N-terminal domain of each NR4A receptor interacts with a unique set of proteins that specifically regulates each of their activities, if it were not for the fact that this review has shown that the interacting partners of the NR4As strongly overlap. However, a closer look at the N-terminal domains of Nur77, Nurr1 and NOR-1 reveals small stretches of relatively high similarity within the amino acid sequences (Fig. 4). The possible importance of these small stretches of high similarity is most readily apparent when looking at phosphorylation sites of the NR4As.
Fig. 4. Amino-acid sequence similarity between the N-terminal domains of the NR4A receptors. The amino-acid sequence of the N-terminal domains of Nur77, Nurr1 and NOR-1 was aligned and the extent of sequence similarity is indicated with colors; e.g. blue indicates the regions where the sequence of the three NR4As is identical. In the Nur77 sequence, the CHEK2 target Thr88, the JNK1 target Ser95, the ERK2 target Thr143, the CK2 target Ser152, and the DNA-PK target Ser164 are indicated with arrows. In the Nurr1 sequence, the ERK2 targets Ser126 and Thr132, and the ERK5 targets Thr168 and Ser177 are indicated with arrows.
7.5.2 The interplay of NR4A receptors and the oncogene–tumor suppressor networks in cancer
Beard JA, Tenga A, Chen T
Cell Signal. 2015 Feb; 27(2):257-66
http://dx.doi.org/10.1016/j.cellsig.2014.11.009
Highlights
- The expression and function of NR4As are dysregulated in multiple cancer types.
- NR4As are positively regulated by oncogenic signaling pathways.
- NR4As are capable of inhibiting tumor suppressor signaling.
- The connectedness of NR4As with these pathways mediate their functions in cancer.
- NR4A agonists and antagonists offer therapeutic strategies for cancer treatment.
Abstract
Nuclear receptor (NR) subfamily 4 group A (NR4A) is a family of three highly homologous orphan nuclear receptors that have multiple physiological and pathological roles, including some in cancer. These NRs are reportedly dysregulated in multiple cancer types, with many studies demonstrating pro-oncogenic roles for NR4A1 (Nur77) and NR4A2 (Nurr1). Additionally, NR4A1 and NR4A3 (Nor-1) are described as tumor suppressors in leukemia. The dysregulation and functions of the NR4A members are due to many factors, including transcriptional regulation, protein-protein interactions, and post-translational modifications. These various levels of intracellular regulation result from the signaling cross-talk of the NR4A members with various signaling pathways, many of which are relevant to cancer and likely explain the family members’ functions in oncogenesis and tumor suppression. In this review, we discuss the multiple functions of the NR4A receptors in cancer and summarize a growing body of scientific literature that describes the interconnectedness of the NR4A receptors with various oncogene and tumor suppressor pathways.
NR4As are positively regulated by oncogenic signaling pathways

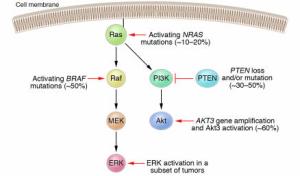
NR4A subfamily of nuclear receptors
http://ars.els-cdn.com/content/image/1-s2.0-S0898656814003556-gr1.sml
intracellular regulation result from the signaling cross-talk of the NR4A members
http://ars.els-cdn.com/content/image/1-s2.0-S0898656814003556-gr2.sml
7.5.3 NLRX1 acts as tumor suppressor by regulating TNF-α induced apoptosis
Singh K, Poteryakhina A, Zheltukhin A, …Chumakov PM, Singh R.
Biochim Biophys Acta. 2015 May; 1853(5):1073-86
http://dx.doi.org/10.1016/j.bbamcr.2015.01.016
Highlights
- NLRX1 sensitizes cancer cells to TNF induced cell death by regulating Caspase-8.
- NLRX1 localizes to mitochondria (mt) and regulates TNF induced mt-ROS generation.
- Mitochondrial association of Caspase-8 with NLRX1 may regulate mt-ETC function.
- NLRX1 expression in cancer cells suppresses tumorigenicity in nude mice.
Chronic inflammation in tumor microenvironment plays an important role at different stages of tumor development. The specific mechanisms of the association and its role in providing a survival advantage to the tumor cells are not well understood. Mitochondria are emerging as a central platform for the assembly of signaling complexes regulating inflammatory pathways, including the activation of type-I IFN and NF-κB. These complexes in turn may affect metabolic functions of mitochondria and promote tumorigenesis. NLRX1, a mitochondrial NOD-like receptor protein, regulate inflammatory pathways, however its role in regulation of cross talk of cell death and metabolism and its implication in tumorigenesis is not well understood. Here we demonstrate that NLRX1 sensitizes cells to TNF-α induced cell death by activating Caspase-8. In the presence of TNF-α, NLRX1 and active subunits of Caspase-8 are preferentially localized to mitochondria and regulate the mitochondrial ROS generation. NLRX1 regulates mitochondrial Complex I and Complex III activities to maintain ATP levels in the presence of TNF-α. The expression of NLRX1 compromises clonogenicity, anchorage-independent growth, migration of cancer cells in vitro and suppresses tumorigenicity in vivo in nude mice. We conclude that NLRX1 acts as a potential tumor suppressor by regulating the TNF-α induced cell death and metabolism.
7.5.4 The Mre11 Complex Suppresses Oncogene-Driven Breast Tumorigenesis and Metastasis
Gupta GP, Vanness K, Barlas A, Manova-Todorova KO, Wen YH, Petrini JH
Mol Cell. 2013 Nov 7;52(3):353-65
http://dx.doi.org/10.1016%2Fj.molcel.2013.09.001
The DNA damage response (DDR) is activated by oncogenic stress, but the mechanisms by which this occurs, and the particular DDR functions that constitute barriers to tumorigenesis, remain unclear. We established a mouse model of sporadic onco-gene-driven breast tumorigenesis in a series of mutant mouse strains with specific DDR deficiencies to reveal a role for the Mre11 complex in the response to oncogene activation. We demonstrate that an Mre11-mediated DDR restrains mammary hyperplasia by effecting an oncogene-induced G2 arrest. Impairment of Mre11 complex functions promotes the progression of mammary hyperplasias into invasive and metastatic breast cancers, which are often associated with secondary inactivation of the Ink4a-Arf (CDKN2a) locus. These findings provide insight into the mechanism of DDR engagement by activated oncogenes and highlight genetic interactions between the DDR and Ink4a-Arf pathways in suppression of oncogene-driven tumorigenesis and metastasis.
The DNA damage response (DDR) network comprises DNA repair, DNA damage signaling, apoptosis, and cell-cycle checkpoint functions (Ciccia and Elledge, 2010). Two lines of evidence support the view that the DDR is a barrier to tumorigenesis. Mutations affecting components of the DDR are frequently associated with predisposition to cancer (Ciccia and Elledge, 2010). Also, indices of DDR activation are evident in preneoplastic lesions or in cultured cells harboring activated oncogenes (Bart-kova et al., 2005; Gorgoulis et al., 2005). Despite supportive genetic data from in vitro and tumor inoculation studies (Bartkova et al., 2006;Di Micco et al., 2006), causal demonstration that the oncogene-induced DDR suppresses tumorigenesis within a tissue context remains limited (Gorrini et al., 2007; Squatrito et al., 2010; Takacova et al., 2012). In certain contexts, the role for ataxia telangiectasia mutated (ATM) in suppressing onco-gene-driven tumorigenesis was relatively minor, although these mouse models were limited by the fact that ATM−/− mice are prone to early spontaneous lymphomagenesis (Efeyan et al., 2009).
The mechanism for DDR activation in response to oncogene expression remains incompletely understood, but the prevailing view posits that oncogene activation leads to replication stress in the form of stalled, and subsequently collapsed, DNA replication forks (Halazonetis et al., 2008). Analysis of the ATRSeckel mouse has indicated that ATR may be required for cell viability upon oncogene activation, suggesting that DNA replication stress may indeed underlie these effects of oncogene activation (López-Contreras et al., 2012;Murga et al., 2011; Schoppy et al., 2012). However, since ATR promotes viability, rather than elimination of the oncogene-expressing cells, this outcome is not consistent with a barrier function for that component of the DDR. The purpose of this study was to delineate the particular aspects of the DDR network that constitute barriers to oncogenesis using a mouse model of sporadic, oncogene-driven breast cancer.
The Mre11 complex is a sensor of DNA double-strand breaks (Stracker and Petrini, 2011). Hypomorphic mutations in this complex, modeled in the mouse after alleles inherited in ataxiatelangiectasia-like disorder (A-TLD) and Nijmegen breakage syndrome (NBS), have facilitated the elucidation of the Mre11 complex’s role in the ATM-dependent DDR. Here, we utilize these and other mutant mouse strains, individually and in combination, to define the tumor-suppressive functions of the DDR in mammary epithelium.
A Mouse Model of Sporadic, Oncogene-Induced Mammary Neoplasia
Expression of activated NeuT (Bargmann and Weinberg, 1988), the rodent ortholog of the ERBB2/HER2oncogene, in the mammary epithelium of adult mice via the RCAS/MMTVTVA system (Du et al., 2006) results in early DDR activation, and oligoclonal tumors with an average latency of 5 months (Reddy et al., 2010). To delineate the aspects of the DDR primarily relevant for tumor suppression in the face of oncogene activation, we interbred MMTV-TVA mice with a variety of mutant mouse strains with established DDR deficiencies. Age-matched cohorts of female animals (12–18 weeks old) were injected with either RCAS-HA-NeuT or control virus via mammary intraductal injection. The genotypes analyzed wereMre11ATLD1/ATLD1, Nbs1ΔB/ΔB, Chk2−/−, Nbs1ΔC/ΔC Chk2−/−, p53515C/515C, p53−/−, and 53BP1−/−, each of which exhibits defects in DNA-damage-induced cell-cycle checkpoint activation, apoptosis, and/or DNA repair (Figures S1A and S1B available online; Liu et al., 2004; Shibata et al., 2010; Stracker et al., 2007, 2008; Stracker and Petrini, 2011; Theunissen et al., 2003; Williams et al., 2002). These mouse strains did not exhibit any histopathological deficits in mammary gland development (data not shown), circumventing the potential problem of differences in mammary tissue among the various genetic backgrounds confounding the analyses.
We performed digital quantification of glandular structures relative to total cellular content in the oncogene-expressing mammary glands and normalized this value to the glandular content observed in the matched control mammary glands (Figure 1C). These variations in mammary ductal enlargement, luminal filling, cellular turnover, and glandular density across the different genotypes are summarized in Figure 1D.
NeuT expression in Chk2−/− and Nbs1ΔC/ΔC Chk2−/− mammary epithelium produced hyperplasias that were only modestly dissimilar from WT (Figures 1B–1D; data not shown), suggesting that apoptosis and the intra-S phase checkpoint—diminished in both mutants (Stracker et al., 2008)—do not mediate the early response to oncogene activation. Consistent with that interpretation, p53515C/515C mutants, in which p53-dependent apoptosis is lost (Liu et al., 2004), also exhibited relatively modest hyper-plasia, although some morphological changes were noted (Figures 1B–1D). In contrast, p53−/− mammary glands resembled p53515C/515C morphologically, but exhibited more extensive NeuT-induced hyperplasia (Figures 1B–1D), consistent with additional deficiencies of the null mutant—including, but not limited to, induction of the G1/S checkpoint and senescence pathways.
In contrast to the aforementioned genotypes, oncogene-induced hyperplasia was markedly distinct in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mammary glands relative to WT mammary glands (Figures 1B–1D). The Mre11 complex mutant genotypes exhibited florid hyperplasia in response to oncogene expression that frequently filled the lumen of the enlarged mammary ducts. Quantification of hyperplasia across the entire mammary gland revealed that Mre11ATLD1/ATLD1 was associated with the most significant degree of oncogene-induced proliferative change (Figure 1C).
We examined oncogene-dependent activation of the DDR in WT and Mre11ATLD1/ATLD1 mammary hyperplasias. Consistent with prior reports (Reddy et al., 2010), we observed the formation of γH2AX foci and accumulation of 53BP1 nuclear staining in WT hyperplasias after the introduction of NeuT (Figures 2A and 2B). We observed a highly significant, >2-fold reduction in both NeuT-induced γH2AX foci formation and 53BP1 accumulation within Mre11ATLD1/ATLD1 lesions relative to WT (p < 0.0001; Figures 2A and 2B). In contrast to the effects of Mre11 complex hypomorphism, oncogene-dependent DDR activation was unperturbed in p53−/− mammary glands (Figure 2A; data not shown). These data demonstrate that the Mre11 complex is required for DDR activation upon NeuT expression.
The oncogene-driven, Mre11 complex-dependent DDR exhibited dissimilarities from that induced by ionizing radiation (IR). First, oncogene expression in the WT mammary gland resulted in finely punctate 53BP1 staining and did not induce the large foci that develop after irradiation of the mammary gland (Figure S4). In addition, phosphorylation of the ATM target KAP1 at Ser824 was not observed in the oncogene-expressing mammary gland, but was readily detected in IR-treated mammary tissue (Figure 2C). Similarly, we observed significantly less p53 stabilization in mammary epithelial cells after oncogene expression in comparison to irradiated tissue (Figure S4). Hence, the Mre11 complex-mediated response to oncogene activation appears to be qualitatively distinct from the response to clastogen-induced DNA damage.
We examined apoptosis and growth arrest—functional outcomes of DDR activation—in hyperplastic lesions. While NeuT expression was associated with increased proliferation and apoptosis rates relative to control mammary glands, we did not observe a statistically significant difference in TUNEL or Ki67 positivity between WT and Mre11ATLD1/ATLD1 oncogene-induced hyperplasias (Figures 3A and 3B). We observed a 4-fold increase in pHH3-S10 staining in WT versus Mre11ATLD1/ATLD1 hyperplasias (p < 0.001; Figure 3C), which was unexpected given the significantly increased cellularity of Mre11ATLD1/ATLD1 hyperplasias. The pHH3-S10 staining pattern that we observed was punctate, and pHH3-S10-positive nuclei did not exhibit morphological features of mitosis (Figure 3C, inset), suggesting that the pHH3-S10 signal represented pericentromeric staining characteristic of late G2 cells rather than mitotic cells.
Centriole duplication was evident in 84% of pHH3-S10-positive cells, compared to only 16% of pHH3-S10-negative cells (p < 0.0001; Figure 4B), indicating a cell-cycle state that is beyond the G1/S transition. These observations collectively suggest that NeuT expression in mammary epithelium activates a Mre11 complex-dependent G2 arrest or accumulation. Notably, this G2 arrest is distinct from the canonical IR-induced G2/M checkpoint, which is also Mre11 dependent (Theunissen et al., 2003). In that context, pHH3-S10 is not induced, suggesting that the heterochromatin-associated accumulation of this marker is oncogene specific.
The variable and prolonged latency of tumor onset in Mre11ATLD1/ATLD1 animals suggests that additional genetic alterations may be required for NeuT-mediated transformation of mammary epithelial cells. We examined p19Arf expression—a well-established oncogene-induced tumor-suppressive pathway (Sherr, 2001)—in the 3-week-old NeuT-expressing mammary hyperplasias from WT and Mre11ATLD1/ATLD1animals. We observed >10-fold induction of p19Arf after oncogene expression in Mre11ATLD1/ATLD1relative to control-injected mammary glands (Figure 6A). The extent of p19Arf induction in NeuT-expressingWT mammary glands was <50% of that observed in Mre11ATLD1/ATLD1 (p < 0.007, Figure 6A). Notably, there was no difference in HA-NeuT expression levels between the WT and Mre11ATLD1/ATLD1 mice that could account for the elevated levels of p19Arf (Figure S6A). As expected, p53 levels were modestly elevated in Mre11ATLD1/ATLD1 hyperplasias relative to WT (Figure S6B).
Collectively, the findings presented here indicate that the Mre11 complex constitutes an inducible barrier to oncogene-driven neoplasia. In response to oncogene activation, the Mre11 complex mediates a G2 arrest that appears to be qualitatively distinct from that revealed in previous analyses of Mre11 complex-dependent DDR functions (Figure 7E; Stracker et al., 2004). The arrest is associated with heterochromatin changes, including the appearance of macroH2A and histone H3 (Ser10) phosphorylation. Histone H3 phosphorylation at pericentric heterochromatin begins early in G2 phase and expands as cells enter mitosis (Crosio et al., 2002). That fact, along with the finding that H3 phosphorylation arises in cells that have undergone centriole duplication, indicates that cells in oncogene-expressing hyperplasias accumulate in G2. We cannot exclude the possibility that other NeuT-expressing cells also arrest in G1 without the observed heterochromatic changes. In Mre11ATLD1/ATLD1 mammary epithelium, the NeuT-induced arrest is lost, and macroH2A and histone H3 phosphorylation are not detected in hyperplastic tissue, demonstrating that the G2 accumulation depends on the Mre11 complex.
The Mre11 complex-dependent G2 arrest does not appear permanent, as WT cells are capable at low frequency of progressing to tumors. When the arrest is attenuated, as in Mre11ATLD1/ATLD1, we observe more extensive oncogene-induced mammary hyperplasia, and a significantly greater likelihood of progression to invasive breast cancer. Although previous studies show that the Mre11 complex suppresses genome instability, and thus the risk of spontaneous DNA-damage-associated tumorigenesis (Stracker et al., 2008; Theunissen et al., 2003), this study demonstrates that the Mre11 complex also suppresses oncogene-driven neoplasia and tumorigenesis.
An important question concerns the underlying basis of the response to oncogene activation. Given the importance of the Mre11 complex in sensing DNA double-strand breaks and initiating an ATM-dependent DDR, a parsimonious interpretation is that oncogene activation results in DNA damage. Indeed, there are compelling genetic data supporting the induction of DNA replication stress upon oncogene activation (Bartkova et al., 2006; Campaner and Amati, 2012; Di Micco et al., 2006; Dominguez-Sola et al., 2007;López-Contreras and Fernandez-Capetillo, 2010). DNA replication stress is a common precursor of frank DNA damage when forks collapse (Allen et al., 2011), which would readily account for the induction of DNA damage upon oncogene induction.
Potential crosstalk between the oncogene-induced DDR and the Arf tumor suppressor pathways has recently been described (Evangelou et al., 2013; Monasor et al., 2013; Velimezi et al., 2013). Our data provide direct evidence for a genetic interaction between these pathways during oncogene-driven tumorigenesis. We demonstrate that when Mre11 complex function is impaired, oncogene expression induces Arf expression, and Ink4a-Arf inactivation is commonly observed in the mammary tumors that ensue. The mechanism for how Mre11 hypomorphism promotes oncogene-induced Arf expression remains unclear. We observe that 40% of the NeuT-induced mammary tumors that developed in Mre11ATLD1/ATLD1 mice had genetic inactivation of the Ink4a-Arf locus, and the remaining tumors exhibited reduced p19Arf expression, suggesting alternative modes of pathway suppression. These findings provide compelling genetic evidence for the cooperative roles of the Mre11 complex and Ink4a-Arf pathways in the suppression of oncogene-driven tumorigenesis and metastasis.
The behavior of the emergent tumors in Mre11ATLD1/ATLD1mice suggests a link between increased chromosomal instability and an elevated rate of metastatic dissemination from the primary tumor. The observation that all of the Ink4a-Arf mutated mammary tumors were lung metastatic also raises the possibility that Arf loss promotes metastatic progression in the context of Mre11 complex impairment.
Our genetic data suggest that functional hypomorphism of this pathway may be a driver of breast tumorigenesis, genomic instability, and metastasis. Given the profound DDR defects associated with Mre11 complex hypomorphism (Stracker and Petrini, 2011), this subset of human breast cancer may exhibit exquisite DNA damage sensitivities that could be therapeutically exploited to improve clinical outcomes.
7.5.5 Expression of Stromal Cell-derived Factor 1 and CXCR4 Ligand Receptor System in Pancreatic Cancer
Koshiba T, Hosotani R, Miyamoto Y, Ida J, …, Fujii N, Imamura M
Clin Cancer Res Sep; 6(9):3530-5
NR4A subfamily of nuclear receptors
http://clincancerres.aacrjournals.org/content/6/9/3530.long
To examine the expression of the stromal cell-derived factor 1 (SDF-1)/CXCR4 receptor ligand system in pancreatic cancer cells and endothelial cells, we performed immunohistochemical analysis for 52 pancreatic cancer tissue samples with anti-CXCR4 antibody and reverse transcription-PCR analysis for CXCR4 and SDF-1 in five pancreatic cancer cell lines (AsPC-1, BxPC-3, CFPAC-1, HPAC, and PANC-1), an endothelial cell line (HUVEC), and eight pancreatic cancer tissues. We then performed cell migration assay on AsPC-1 cells, HUVECs, and CFPAC-1 cells in the presence of SDF-1 or MRC-9 fibroblast cells. Immunoreactive CXCR4 was found mainly in pancreatic cancer cells and endothelial cells of relatively large vessels around a tumorous lesion. The immunopositive ratio in the pancreatic cancer was 71.2%. There was no statistically significant correlation with clinicopathological features. SDF-1 mRNA expressions were detected in all pancreatic cancer tissues but not in pancreatic cancer cell lines and HUVECs; meanwhile, CXCR4 mRNA was detected in all pancreatic cancer tissues, cancer cell lines, and HUVECs. The results indicate that the paracrine mechanism is involved in the SDF-1/CXCR4 receptor ligand system in pancreatic cancer. In vitro studies demonstrated that SDF-1 significantly increased the migration ability of AsPC-1 and HUVECs, and these effects were inhibited by CXCR4 antagonist T22, and that the coculture system with MRC-9 also increased the migration ability of CFPAC-1 cells, and this effect was significantly inhibited by T22. Our results suggested that the SDF-1/CXCR4 receptor ligand system may have a possible role in the pancreatic cancer progression through tumor cell migration and angiogenesis.
Chemokines belong to the small molecule chemoattractive cytokine family and are grouped into CXC chemokines and CC chemokines, on the basis of the characteristic presence of four conserved cysteine residues (1, 2, 3) . Chemokines mediate the chemical effect on target cells through G-protein-coupled receptors, which are characterized structurally by seven transmembrane spanning domains and are involved in the attraction and activation of mononuclear and polymorphonuclear leukocytes. The effects of CXC chemokines on cancer cells have been investigated in the case of IL3 -8. Several studies have demonstrated the presence of IL-8 and its receptor in tumor tissues, which were involved in vascular endothelial cell proliferation and tumor neovascularization ,(4, 5, 6, 7) . It was also reported that IL-8 inhibited non-small cell lung cancer proliferation via the autocrine and paracrine pathway (8) . IL-8 produced by malignant melanoma was found to induce cell proliferation via the autocrine pathway in vitro (9) . These studies indicate that IL-8 is involved in the regulation of tumor progression through tumor angiogenesis and/or direct cancer cell growth.
SDF-1 was initially cloned by Tashiro et al. (10) and later identified as a growth factor for B cell progenitors, a chemotactic factor for T cells and monocytes, and in B-cell lymphopoiesis and bone marrow myelopoiesis (11, 12, 13) . SDF-1 is a member of the CXC subfamily of chemokines, and its chemotactic effect is mediated by the chemokine receptor CXCR4 (12 , 14) . Most of the chemokine receptors interact with pleural ligands, and vice versa, but the SDF-1/CXCR4 receptor ligand system has been shown to involve a one-on-one interaction (15 , 16) . Furthermore, CXCR4 has been shown to function as a coreceptor for T lymphocytotrophic HIV-1 isolates (17) . Recent studies have demonstrated that endothelial cells express CXCR4 and are strongly chemoattracted by SDF-1 (18, 19,20) . Tachibana et al. (15) reported that in the embryo of CXCR4 or SDF-1 knockout mice larger branches of the superior mesenteric artery were missing and that the resultant abnormal circulatory system led to gastrointestinal hemorrhage and intestinal obstruction. These findings suggest that SDF-1 and CXCR4 are involved in organ vascularization, as well as in the immune and hematopoietic system.
To clarify the role of the SDF-1/CXCR4 receptor ligand system in pancreatic cancer, we have investigated the expression of CXCR4 and SDF-1 with the aid of immunohistochemical analysis and RT-PCR in pancreatic cancer tissue and experimental chemotactic activity of SDF-1 in pancreatic cancer cells and vascular endothelial cells in vitro.
The distribution of CXCR4 protein expression in pancreatic cancer tissue was examined by means of immunohistochemical analysis of pancreatic cancer tissue samples obtained at surgical operation. Fig. 1<$REFLINK> shows representative immunostainings of cancerous and noncancerous regions in pancreatic cancer tissues. Staining of the CXCR4 protein was identified in the cytoplasm and/or cell membrane of cancer cells, but was not detected in the normal acinar cells and ductal cells of noncancerous region in pancreatic cancer tissue. Negative or weak staining for the CXCR4 protein was observed in a majority of the infiltrating inflammatory cells in the specimens. The immunopositive ratio of cancer cells in the pancreatic cancer tissue specimens was 71.2% (37 of 52). Table 1<$REFLINK>summarizes the relationship between CXCR4 expression and clinicopathological features of 52 pancreatic cancers. There was no significant correlation between the expression of CXCR4 protein and the clinicopathological variables examined (i.e., tumor extension, lymph node metastasis, liver metastasis, and Union International Contre Cancer stage). CXCR4 immunoreactivities were observed in endothelial cells of relatively large vessels around the tumorous lesions, but were scarcely found in the endothelial cells of microvessels inside tumorous lesions (Fig. 2, A and B)<$REFLINK> .
We performed RT-PCR using specific primers, as described in“ Materials and Methods,” to confirm CXCR4 and SDF-1 mRNA expression in pancreatic cancer cells, endothelial cells (HUVECs), and pancreatic cancer tissues. CXCR4 mRNA expressions were clearly detected in five pancreatic cancer cell lines, HUVECs, and eight pancreatic cancer tissue samples (Fig. 3a)<$REFLINK> . On the other hand, SDF-1 mRNA expression was not detected in five pancreatic cancer cell lines and HUVECs, but was identified in eight pancreatic cancer tissue samples (Fig. 3b)<$REFLINK> .
Transwell migration assays were performed to examine the effects of SDF-1 on motility of pancreatic cancer cells (AsPC-1) and endothelial cells (HUVEC). At a concentration of 100 ng/ml, SDF-1 induced chemotaxis of AsPC-1 cells, which was approximately double that of the control. One micromolar of T22 (CXCR4 antagonist) and 10 μg/ml of IVR7 (neutralizing CXCR4 antibody) completely blocked the chemotaxis of AsPC-1 induced by 100 ng/ml SDF-1 (Fig. 4a)<$REFLINK> . At a concentration of 100 g/ml SDF-1 induced an approximately quadruple chemotaxis of HUVECs. One micromolar of T22 caused a 33% reduction of the chemotaxis of HUVECs in the presence of containing 100 ng/ml SDF-1 (Fig. 4b)<$REFLINK> .
SDF-1 belongs to the CXC chemokine family and is a ligand for CXCR4. The role of the SDF-1/CXCR4 receptor ligand system has been investigated mainly in the field of immunology, especially in the mechanism of infection of T lymphocytotrophic HIV-1 and for the prevention of HIV-1 infection. Investigators have also paid attention to the role of the SDF-1/CXCR4 receptor ligand system in cancer tissues.
In this study, we first used immunohistochemical methods to examine CXCR4 expression in pancreatic cancer tissues. Immunoreactive CXCR4 was found in the cytoplasm and/or cell membrane of pancreatic cancer cells. Although CXCR4 staining in pancreatic cancer tissue was heterogeneous and showed differences between specimens, it was found mainly in cancer cells: the immunopositive ratio for the pancreatic cancer tissue specimens was 71.2% (37 of 52). There was a tendency for the immunopositive ratio of CXCR4 in tumors with lymph node metastasis or liver metastasis to be higher than in tumors without these features, but no statistically significant correlation with clinicopathological features were found. There is a diversity of views on the role of the SDF-1/CXCR4 receptor ligand system in malignant tissues. In the current study, SDF-1 mRNA expressions were detected in all pancreatic cancer tissues (eight of eight) but were not detected in pancreatic cancer cell lines (zero of five), whereas CXCR4 mRNA was detected in both pancreatic cancer tissues (eight of eight) and cancer cell lines (five of five). The results indicate that the paracrine mechanism may be involved in the SDF-1/CXCR4 receptor ligand system in pancreatic cancer.
Our results suggest that the SDF-1/CXCR4 receptor ligand system may have a possible role in the pancreatic cancer progression through tumor cell migration and angiogenesis. Because T22 suppressed the migration of both pancreatic cancer cells and endothelial cells in vitro, additional in vivo studies are warranted to examine whether T22 suppresses the tumor spread and tumor angiogenesis to clarify the role of the SDF-1/CXCR4 receptor ligand system in pancreatic cancer.
7.5.6 DLC1- a significant GAP in the cancer genome
Aurelia Lahoz and Alan Hall
Genes Dev. 2008 Jul 1; 22(13): 1724–1730
http://dx.doi.org/10.1101.2Fgad.1691408
Rho GTPases are believed to make important contributions to the development and progression of human cancer, but direct evidence in the form of somatic mutations analogous to those affecting Ras has been lacking. A recent study in Genes & Development by Xue and colleagues (1439–1444) now provides in vivo evidence that DLC1, a negative regulator of Rho, is a tumor suppressor gene deleted almost as frequently as p53 in common cancers such as breast, colon, and lung.
Cancer is a complex set of diseases arising from combinations of genetic and epigenetic events, including base mutations, chromosomal rearrangements, DNA methylation, and chromatin modification. Genetic changes were first seen cytologically and revealed gross chromosomal abnormalities, such as translocations, deletions, amplifications (of entire chromosomes or parts of chromosomes), and inversions. Subsequently, DNA sequencing of candidate genes and then whole genomes has uncovered large numbers of more subtle genetic alterations. The recent and continuing successes of sequencing and other nonfunctional based genomic approaches have raised new problems in how to determine which changes have significance for tumor development. This is not a trivial problem and will require combinations of cell-based assays, in vivo animal models, and ultimately clinical intervention.
The identification of the Ras oncogene was the first major triumph of the early application of molecular biology to the cancer problem (Malumbres and Barbacid 2003). Although originally identified as a viral oncogene in a rodent sarcoma-inducing retrovirus, it was the seminal work of the Weinberg and Cooper laboratories in 1981 (Krontiris and Cooper 1981; Shih et al. 1981), using DNA transfection assays of human tumor DNA into immortalized mouse fibroblasts, that led to the identification of Ras as a true human oncogene. Several groups went on to show that any one of the three Ras genes (HRAS, KRAS, and NRAS) could be converted into a human oncogene by a single base mutation leading to a single amino acid substitution in the encoded Ras protein. Ras mutations are found in ∼30% of most, though not all, cancer types and it remains the most frequently mutated dominant oncogene so far identified (Bos 1989). We now know much about the consequences of those amino acid substitutions and the cellular and physiological importance of Ras in controlling proliferation and differentiation. Ras is an example of a regulatory GTPase that cycles between active (GTP-bound) and inactive (GDP-bound) conformations to control biochemical pathways and processes. These molecular switches are activated by guanine nucleotide exchange factors (GEFs), which catalyze exchange of GDP for GTP, and are inactivated by GTPase-activating proteins (GAPs), which promote the otherwise slow, intrinsic GTPase activity of the proteins (Fig. 1). The amino acid substitutions identified in Ras in human cancers are found at codons 12, 61, and to a lesser extent 13, and the common consequence of these changes is to prevent GAP-mediated stimulation of GTP hydrolysis leading to permanent activation of the switch (Trahey and McCormick 1987). Inspection of Figure 1 suggests possible alternative ways in which this molecular switch could be inappropriately activated. For example, activating mutations in one of the nine RasGEF genes or inactivation of one of the eight RasGAP genes could lead to hyperactivation of the switch. To date, no such mutations have been reported in GEF genes in human cancers, but one of the GAPs, neurofibromin, is encoded by the NF1 tumor suppressor gene. Patients with neurofibromatosis type I inherit only one functional NF1 gene and are then predisposed to cancer through complete loss of NF1. In addition, mutational activation of components of downstream signaling pathways (Fig. 1) could bypass the need for Ras and this is clearly the case with somatic mutations in BRAF (which encodes a Ras effector), found most frequently in malignant melanomas (>50%), but also in thyroid, colorectal, and ovarian cancer (Davies et al. 2002; Wellbrock et al. 2004).

The Ras GTP.GDP cycle
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2732422/bin/1724fig1.jpg
Figure 1. The Ras GTP/GDP cycle. Ras GTPases are molecular switches and the GDP/GTP cycle is controlled by GEFs and GAPs. The output of the switch is through the interaction of Ras.GTP with effector proteins.
Rho GTPases can trigger numerous downstream signaling pathways by interacting with distinct effectors—to date, ∼20 such target proteins have been reported that specifically interact with Rho (Etienne-Manneville and Hall 2002). One of the best-characterized is Rho kinase (ROCK), which regulates myosin II and actin filament contractility, through its ability to phosphorylate and inactivate myosin light chain phosphatase (Fukata et al. 2001). Rho kinase is involved in many aspects of normal cell biology, such as cell cycle, morphogenesis, and migration, and in addition has been shown to participate in the proliferation, invasion, and metastasis of cancer cells (Etienne-Manneville and Hall 2002; Sahai and Marshall 2002; Narumiya and Yasuda 2006). In the final part of their study, Xue et al. (2008) show that two small molecule Rho kinase inhibitors, Y-27632 and to a lesser extent Fasudil, inhibit in vitro colony formation of p53−/− liver progenitor cells expressing c-Myc and DCL1 shRNA. It should be noted, however, that both Y-27632 and Fasudil inhibit PRK/PKN and citron kinase, two other kinases activated by Rho, so the result is not entirely conclusive (Ishizaki et al. 2000).
Embryonic fibroblasts can be obtained from DLC1−/− mice and these display alterations in the organization of actin filaments and focal adhesion (Durkin et al. 2005). Confusingly, however, these knockout cells have fewer stress fibers and focal adhesions—the opposite of what would have been predicted for the loss of a GAP that regulates Rho. In fact the cytoskeletal and adhesion complex changes seen in DLC−/− fibroblasts appear to be more in keeping with Rac activation. Unfortunately the authors did not examine the levels of either Rho.GTP or Rac.GTP in these cells, which might have provided some insight into this unexpected result. In the absence of tissue-specific mouse knockouts, we must look to work in Drosophila on RhoGAP88C, the fly ortholog of DCL1, to provide some in vivo physiological data. Mutations in RhoGAP88C were first identified as crossveinless-c and result in defects in tissue morphogenesis during development (Denholm et al. 2005). Closer examination suggests that this GAP regulates tubulogenesis and convergent extension, two processes driven by reorganization of the actin cytoskeleton. An additional and provocative observation to emerge from this study is that RhoGAP88C acts through Rho in some tissues, but it acts through Rac and not Rho in others. The in vitro biochemical activity of this GAP has not been determined and so it is possible that it shows a different specificity from its mammalian counterpart. Otherwise, tissue-specific modification of its catalytic activity would need to be invoked, rendering the in vitro assays essentially useless for predicting specificity. Two subsequent studies have concluded that RhoGAP88C is localized basolaterally in epithelial cells and serves to restrict Rho activity to the apical surface and thereby generate morphogenetic tissue remodeling through polarized activation of myosin II (Brodu and Casanova 2006; Simoes et al. 2006).
Taken together, a picture emerges of spatially localized DLC1 acting to control Rho activity so as to promote changes in the actin cytoskeleton during cell morphogenesis. The disruption of this pathway might be expected to lead to tissue disorganization during differentiation programs, which could promote inappropriate cell proliferation (Fig. 2).

DLC1 is a tumor suppressor.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2732422/bin/1724fig2.jpg
Figure 2. DLC1 is a tumor suppressor. Loss of DLC1 leads to deregulated and/or delocalized activation of Rho. This may disrupt tissue morphogenesis leading to inappropriate proliferation. (PM) Plasma membrane.
Directed therapeutic intervention depends on a deep understanding of the relevant signaling pathways through which DLC1 loss is manifest. It is a sobering thought that the signaling pathways downstream from Ras responsible for human cancer are still debated some 25 years after its discovery as a human oncogene and it would be optimistic to believe that identifying Rho pathways will be any easier. Inhibiting the GTPase itself, whether Ras or Rho, is challenging. One of the most promising potential targets for Ras inactivation has been farnesyltransferase (FT), the enzyme required for carboxy-terminal, post-translational modification by a farnesyl lipid (Wright and Philips 2006). FT inhibitors are currently in clinical trials, though the data reported so far are not encouraging. Inhibiting Rho using a similar strategy seems less attractive, since it uses a geranylgeranyltransferase to add a geranylgeranyl group; a much more widespread modification than farnesyl addition. Two other processing enzymes that act on both Ras and Rho, a carboxyl-protease and an isoprenylcysteine carboxyl methyltransferase, are being considered as Ras targets, but in tissue culture at least these seem not to be essential for Rho function (Michaelson et al. 2005). Another possibility that is distinctive to DLC1 might be to attack the epigenetic mechanisms that appear to be commonly used to silence this gene in human cancers. Inhibitors of DNA methyltransferase and histone deacetylase (HDAC) have already been shown to induce the restoration of DLC1 expression in cancer cells, making Zebularine, a new and highly effective DNA demethylating agent, as well as HDAC inhibitors attractive therapeutic approaches (Guan et al. 2006; Neureiter et al. 2007; Seng et al. 2007; Xu et al. 2007). Finally, if it turns out that Rho kinase mediates the key signaling pathway downstream from DLC1 loss, then there is already a huge effort underway to develop small molecule inhibitors of this protein. Rho kinase has been implicated in various forms of cardiovascular disease—such as pulmonary hypertension, myocardial hypertrophy, and atherosclerosis—and in fact one compound, Fasudil, is already being used clinically in Japan for cerebral ischemia (Rikitake and Liao 2005; Tawara and Shimokawa 2007). With over a dozen pharmaceutical companies reportedly working on this problem, and if the work from Xue et al. (2008) implicating Rho kinase downstream from DLC1 turns out to be correct, those companies may end up with a blockbuster!
7.5.7 DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma.
Xue W, Krasnitz A, Lucito R, Sordella R, … , Zender L, Lowe SW.
Genes Dev. 2008 Jun 1;22(11):1439-44
http://dx.doi.org/10.1101.2Fgad.1672608
Deletions on chromosome 8p are common in human tumors, suggesting that one or more tumor suppressor genes reside in this region. Deleted in Liver Cancer 1 (DLC1) encodes a Rho-GTPase activating protein and is a candidate 8p tumor suppressor. We show that DLC1 knockdown cooperates with Myc to promote hepatocellular carcinoma in mice, and that reintroduction of wild-type DLC1 into hepatoma cells with low DLC1 levels suppresses tumor growth in situ. Cells with reduced DLC1 protein contain increased GTP-bound RhoA, and enforced expression a constitutively activated RhoA allele mimics DLC1 loss in promoting hepatocellular carcinogenesis. Conversely, down-regulation of RhoA selectively inhibits tumor growth of hepatoma cells with disabled DLC1. Our data validate DLC1 as a potent tumor suppressor gene and suggest that its loss creates a dependence on the RhoA pathway that may be targeted therapeutically.
Tumor suppressor genes act in signaling networks that protect against tumor initiation and progression, and can be inactivated by deletions, point mutations, or promoter hypermethylation. Although tumor suppressors are rarely considered direct drug targets, they can negatively regulate pro-oncogenic signaling proteins that are amenable to small molecule inhibition. For instance, NF1 inhibits the Ras signaling pathway, which is deregulated in many cancers and has been pursued for its therapeutic potential (Downward 2003). Similarly, PTEN inhibits the PI3–kinase pathway, and inhibitors of PI3K pathway components such as PI3K, AKT, and mTORs have entered clinic trials (Luo et al. 2003).
Recurrent chromosomal deletions found in sporadic cancers often contain tumor suppressor genes. For example, PTEN loss on chromosome 10q23 frequently occurs in various cancers and promotes tumorigenesis by deregulating the PI3 kinase pathway (Maser et al. 2007). Similarly, heterozygous deletions on chromosome 8p22 in many hepatocellular carcinomas (HCC) (Jou et al. 2004) and other cancer types, including carcinomas of the breast, prostate, colon, and lung (Matsuyama et al. 2001; Durkin et al. 2007). Several genes, including DLC1, MTUS1, FGL1 and TUSC3, have been identified as candidate tumor suppressors in this region (Yan et al. 2004). Deleted in Liver Cancer 1 (DLC1) is a particularly attractive candidate owing to its genomic deletion, promoter methylation, and underexpressed mRNA in cancer (Yuan et al. 1998, 2003a; Ng et al. 2000; Wong et al. 2003; Guan et al. 2006; Seng et al. 2007; Ying et al. 2007;Zhang et al. 2007; Pike et al. 2008; for review, see Durkin et al. 2007).
Despite its potential importance, functional data implicating DLC1 loss in tumorigenesis are lacking. DLC1encodes a RhoGAP protein that catalyzes the conversion of active GTP-bound RhoGTPase (Rho) to the inactive GDP-bound form and thus suppresses Rho activity (Yuan et al. 1998). DLC1 has potent GAP activity for RhoA and limited activity for CDC42 (Wong et al. 2003; Healy et al. 2008). When overexpressed, DLC1 inhibits the growth of tumor cells and xenografts (Yuan et al. 2003b, 2004; Zhou et al. 2004; Wong et al. 2005; Kim et al. 2007), but whether this requires its Rho-GAP activity or other functions remains unresolved (Qian et al. 2007; Liao et al. 2007). Most functional studies to date have relied on DLC1 overexpression and, as yet, none have documented that loss of DLC1 promotes transformation in vitro or tumorigenesis in vivo. Indeed, homozygous dlc1 knockout mice die around embryonic day 10.5 (E10.5), and there is no overt phenotype in dlc1 heterozygous mice (Durkin et al. 2005).
Our laboratory recently developed a “mosaic” mouse model whereby liver carcinomas can be rapidly produced with different genetic alterations by manipulation of cultured embryonic liver progenitor cells (hepatoblasts) followed by transplantation into the livers of recipient mice (Zender et al. 2005, 2006). We previously used this model to identify new oncogenes in HCC, which could be characterized in an appropriate biological and genetic context (Zender et al. 2006). Furthermore, using this system, we showed that shRNAs capable of suppressing gene function by RNAi could recapitulate the consequences of tumor suppressor gene loss on liver carcinogenesis (Zender et al. 2005; Xue et al. 2007). Here we combine this mosaic model and RNAi to validate DLC1 as a potent tumor suppressor gene and study its action in vivo.
Studies using low-resolution genome scanning methods have identified chromosome 8p deletions as common lesions in liver carcinoma and other tumor types. To confirm and extend these observations, we examined a series of data sets of copy number alterations in HCC obtained using representational oligonucleotide microarray analysis (ROMA), a variation of array-based CGH that enables genome scanning at high resolution (Lucito et al. 2003). In a panel of 86 liver cancers, heterozygous deletions encompassing theDLC1 were observed in 59 tumors (Fig. 1A,B; data not shown). Consistent with previous reports, these deletions were large (>5 Mb), encompassing >20 annotated genes but invariably included the DLC1 locus. Indeed, heterozygous deletions of DLC1 occurred more frequently than those observed for the well-established tumor suppressors such as INK4a/ARF, PTEN, and TP53 (Fig. 1C). Furthermore, DLC1deletions were nearly as common as those for TP53 in other major tumor types such as lung, colon, and breast (Fig. 1C). Again, most 8p deletions were large, although in breast cancer DLC1 resided at a local deletion epicenter reminiscent of that surrounding the INK4a/ARF locus on chromosome 9p21 (Fig. 1D,E). Although we did not examine the status of the remaining allele in this tumor cohort, studies suggest that it can be silenced by promoter methylation (Yuan et al. 2003a; for review, see Durkin et al. 2007). Together, these data suggest that DLC1 loss plays an important role in human cancer but, in the absence of functional validation, are not conclusive.
Genetically modified liver progenitors were seeded into the livers of syngeneic recipients to assess their ability to form tumors in situ. In contrast to the modest impact of DLC1 loss in vitro, DLC1 shRNAs significantly accelerated tumor onset in vivo (P value < 0.0001 for shDLC1-1 and P < 0.0005 for shDLC1-2) (Fig. 2D,E). In fact, at 57 d post-transplantation, GFP-positive tumor nodules were observed in the livers of most animals receiving cells harboring DLC1 shRNAs, whereas the control animals showed no macroscopically detectable tumor burden (Fig. 2E). Furthermore, the pathology of tumors derived from DLC1 knockdown resembled aggressive human HCC and displayed a high proliferative index as assessed by Ki67 immunohistochemistry (Fig. 2F). Tumors also expressed the HCC markers α-fetoprotein (AFP) and albumin (Supplemental Fig. S3B). These data demonstrate that loss of DLC1 can efficiently promote the development of HCC.
We also ectopically expressed the murine dlc1 gene in mouse hepatoma cells and tested their ability to form tumors orthotopically. To this end, we cloned a Myc-tagged murine dlc1 cDNA and confirmed its ability to produce a protein of the correct molecular weight (Fig. 3A). A mouse hepatoma cell line harboring a luciferase reporter and expressing oncogenic Ras and undetectable DLC1 (see Fig. 1F, lane 8) was infected with the DLC1-expressing retrovirus or an empty vector. Consistent with the literature (Ng et al. 2000), reintroduction of DLC1 produced a modest effect on proliferation in colony formation assays (Supplemental Fig. S4A,B).
Although RhoA has been identified as a DLC1 effector, overexpression studies suggest that other DLC1 functions can contribute to its anti-proliferative activities (Liao et al. 2007; Qian et al. 2007). To determine whether RhoA is required for maintaining tumorigenesis stimulated by DLC1 loss, we tested whether suppression of RhoA in DLC1-suppressed hepatoma lines would impact their expansion as subcutaneous tumors in immunocompromised mice. shRNAs capable of down-regulating RhoA to varying degrees (Fig. 5A) decreased the in vivo growth of two independent murine hepatoma lines with undetectable DLC1 (Fig. 5B, cell lines 1,2; Supplemental Fig. S6A,B). Of note, none of the shRNAs completely suppressed RhoA expression, and their ability to limit tumor expansion was proportional to their knockdown efficiency (Supplemental Fig. S6A). The impact of these shRNAs was less pronounced in hepatoma cell lines with higher DLC1 levels (Fig. 5B, cell lines 3,4; Supplemental Fig. S6C,D). Although complete inhibition of RhoA activity might be generally cytostatic (see Piekny et al. 2005), these data suggest that RhoA is required for maintaining the growth of tumors with attenuated DLC1 activity.
In this study, we combined in vivo RNAi and a mosaic mouse model of HCC to study the impact of DLC1 loss on liver carcinogenesis in mice, which to date has not been possible owing to the embryonic lethality of DLC1 knockout animals. We show that DLC1 loss, when combined with other oncogenic lesions, promotes HCC in vivo and that RhoA activation is both necessary and sufficient for its effects. In our survey of copy number alterations in human tumors, 8p22 deletions encompassing DLC1 occurred in >60% of heptocellular carcinomas as well as a large portion of human lung, breast, and colon carcinomas (see also Durkin et al. 2007). Similarly, RhoA is up-regulated in HCC and many other tumor types (Sahai and Marshall 2002;Fukui et al. 2006). Although other tumor suppressor genes may also reside in the 8p region, our results demonstrate that DLC1 is functionally important and highlight the potential importance of the RhoA signaling network in epithelial cancers.
Molecularly targeted therapies have been devised for inhibiting several oncogenic pathways, including those affected by BCR-ABL, activated Ras and PI3kinase (Downward 2003; Luo et al. 2003). Although tumor suppressors are generally not amenable to direct therapeutic targeting, their mutation may confer a cellular dependency on downstream oncogenic proteins that can be inhibited with small molecule drugs. In this regard, the impact of DLC1 loss may parallel that produced by loss of PTEN, which deregulates the PI3K pathway and can sensitize cells to pharmacological inhibitors of downstream effectors such as mTOR (Maser et al. 2007). Our data indicate that RhoA is required for maintaining at least some tumors driven by DLC1 loss, and that cells with disabled DLC1 are particularly sensitive to inhibitors that target at least one RhoA effector. Clearly, more studies will be required to confirm and extend these observations; nevertheless, the high frequency of DLC1 loss in human cancer implies that pharmacologic intervention of the signaling pathways modulated by DLC1 may have broad therapeutic utility.
7.5.8 Smad7 regulates compensatory hepatocyte proliferation in damaged mouse liver and positively relates to better clinical outcome in human hepatocellular carcinoma
Feng T, Dzieran J, Gu X, Marhenke S, Vogel A, …, Dooley S, Meindl-Beinker NM.
Clin Sci (Lond). 2015 Jun 1; 128(11):761-74
http://dx.doi.org:/10.1042/CS20140606
Transforming growth factor β (TGF-β) is cytostatic towards damage-induced compensatory hepatocyte proliferation. This function is frequently lost during hepatocarcinogenesis, thereby switching the TGF-β role from tumour suppressor to tumour promoter. In the present study, we investigate Smad7 overexpression as a pathophysiological mechanism for cytostatic TGF-β inhibition in liver damage and hepatocellular carcinoma (HCC). Transgenic hepatocyte-specific Smad7 overexpression in damaged liver of fumarylacetoacetate hydrolase (FAH)-deficient mice increased compensatory proliferation of hepatocytes. Similarly, modulation of Smad7 expression changed the sensitivity of Huh7, FLC-4, HLE and HLF HCC cell lines for cytostatic TGF-β effects. In our cohort of 140 HCC patients, Smad7 transcripts were elevated in 41.4% of HCC samples as compared with adjacent tissue, with significant positive correlation to tumour size, whereas low Smad7 expression levels were significantly associated with worse clinical outcome. Univariate and multivariate analyses indicate Smad7 levels as an independent predictor for overall (P<0.001) and disease-free survival (P=0.0123). Delineating a mechanism for Smad7 transcriptional regulation in HCC, we identified cold-shock Y-box protein-1 (YB-1), a multifunctional transcription factor. YB-1 RNAi reduced TGF-β-induced and endogenous Smad7 expression in Huh7 and FLC-4 cells respectively. YB-1 and Smad7 mRNA expression levels correlated positively (P<0.0001). Furthermore, nuclear co-localization of Smad7 and YB-1 proteins was present in cancer cells of those patients. In summary, the present study provides a YB-1/Smad7-mediated mechanism that interferes with anti-proliferative/tumour-suppressive TGF-β actions in a subgroup of HCC cells that may facilitate aspects of tumour progression.
Like this:
Like Loading...
Read Full Post »



 Article Info
Article Info