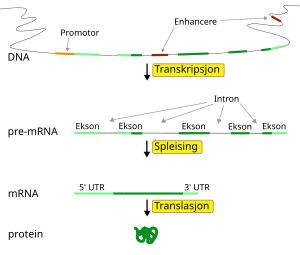
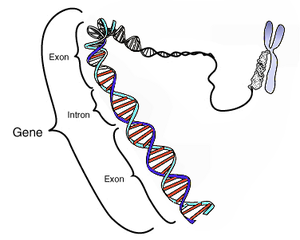
Epigenetic mechanisms

Word Cloud By Danielle Smolyar
The Underappreciated EpiGenome
Author: Demet Sag, PhD
Early 1990’s Kavai group developed a method called Restriction Landmark Genomic Scanning using Methylation-sensitive endonucleases (RLGS-M) to identify differential methylation during development based on CpG islands. In their study they showed that the appearance and disappearance of the spots were specific to tissue and affecting gene regulation.
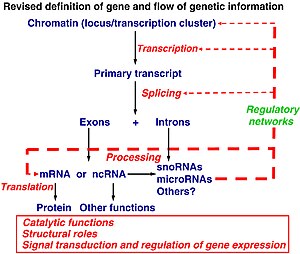
Revised definition of gene and flow of genetic information (adapted from Mattick JS (2003). Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. BioEssays 25:930. doi:10.1002/bies.10332).
Epigenetics is getting a big attention recently to understand genomics and provide better results. However, this field is studied for many years under functional genomics and developmental biology for cellular and molecular biology. Stem cells have a free drive that we have not figured out yet. So genomics must be studied essentially with people training in developmental biology and comparative molecular genetics knowledge to make heads and tail for translational medicine.
There are three main routes of epigenetic modifications one
In 1993, Kavai group showed brain development assays of mice showed that only 0.7% genome has tissue and cellular specificity, and 1.7% of genome was able to turn on and off. This conclusion is relevant to genome sequencing data. Also, previous studies in genome and RNA biology presented that RNA directed DNA modifications lead into splicing and transcriptional silencing for gene regulation in Arapsidosis, mice, and Drosophila. (Borge, F. and. Martiensse, R.A. 2013; Di Croce L, Raker VA, Corsaro M, et al. 2002; Piferrer, F, 2013; Jun Kawai1 et al. 1993)
Comparative developmental biology studies and genetics in organisms give away clues that can be applied or open the door for a new discovery in human disease models. Plants do utilize methylation and transposomes, which are viral particles that can affect the genome structure and present in all organisms including human, for their gene regulation and development extensively (The EMBO Journal, (22 March 2013) | doi:10.1038/emboj.2013.49). Thus, Arapsidosis is a good model.
The environment and gene expression define inheritable materials at transcriptional levels. He group (Cui-Jun Zhang et al, 2013, doi:10.1038/emboj.2013.49) suggested splicing machinery affecting RdDM and transcriptional silencing to control gene regulation during development since an RNA-directed DNA methylation (RdDM) pathway directs de novo methylation. Furthermore, these differences are highly regulated at RNA level with silencing, splicing, transposon activation/inactivation and modulation at epigenetic level.
In fruit fly with more than five hundreds years of accumulated data, the sex determination pathways, soma or germline, share common genes but they act differently in each pathway. Sxl (sex-lethal), which is key gene of somatic sex determination and is an RNA binding protein, regulates the genes through splicing in one its mechanisms (DOI: 10.1002/dvdy.23924). Yet, in germline ovo, which is a DNA binding protein, regulates the expression and development with different sets of rules yet these two paths always communicate to make the final outcome. The effects of RNA on epigenetics and gene regulation during development will be another topic to discuss. However, the three major epigenetic factors do not run in order necessarily, but always have three targeted outcome: initiate, differentiate and maintain. Thus, from environment to phenotype there are places/parts scientists can modulate or reprogram but there parts must be kept intact.
Yang et al, 2012 presented a study on PHD Finger Protein 7 (PHF7), which is an important factor for male germline sexual identity in Drosophila, and called this gene as a “epigenetic reader’ since the expression of this gene in XX soma started a female germline development. This type of epigenetic readers may also alter the outcome to balance cell metabolism towards desired phenotype in stem cells.

Adenine methylation (Photo credit: Allen Gathman)
The environment creates the epigenerators including temperature, differentiation signals and metabolites that trigger the cell membrane proteins for development of signal transduction within the cell to activate gene(s) and to create cellular response. These changes can be modulated but they are not necessary for modulation. The second step involves epigenetic initiators that require precise coordination to recognize specific sequences on a chromatin in response to epigenerator signals. These molecules are
After they are involved they are on for life and controlled by autoregulatory mechanisms, like Sxl (sex lethal) RNA binding protein in somatic sex determination and ovo DNA binding protein in germline sex determination of fruit fly. Both have autoregulation mechanisms, cross talks, differential signals and cross reacting genes since after the final update made the soma has to maintain the decision to stay healthy and develop correctly. Then, this brings the third level mechanism called epigenetic maintainers that are DNA methylating enzymes, histone modifying enzymes and histone variants. The good news is they can be reversed. As a result the phonotype establishes either a
- short term phenotype, transient for transcription,
- DNA replication and repair or
- long term phenotype outcomes that are chromatin conformation and heritable markers.
Early in development things are short term and stop after the development seized but be able to maintain the short term phonotype during wound healing, coagulation, trauma, disease and immune responses. Some cells will loose their ability to differentiate to very low levels. Yet, in life everything is possible even with less than 1% chance because nothing is accidental.
X-chromosome has fascinating characteristics simply because they present unusual mechanisms among female and male differentiation in fruit flies and mammals but their distinct characteristics in evolution marry with surprising parallel mechanisms in regulation. These features are the importance of noncoding RNAs, and epigenetic spreading of chromatin-modifying activities, and at the end of the actions most part of the Y chromosome is lost and one of the X-chromosome is downregulated in the big picture.

DNA methylation analysis methods not based on methylation-specific PCR. Following bisulfite conversion, the genomic DNA is amplified with PCR that does not discriminate between methylated and non-methylated sequences. The numerous methods available are then used to make the discrimination based on the changes within the amplicon as a result of bisulfite conversion. (Photo credit: Wikipedia)
Revisiting RNA directed DNA methylation study once again shows that unread sequence has the word on gene expression; it can still create the diversity that may help rebirth of stem cells with a correct program and develop tools for unmet human diseases. This will be the next topic to discuss.
Personal Impression
While I was listening Dr. Ecker, I remembered these studies. The question becomes what we know then what we know now. “The Underappreciated Epigenome: Methylation of Brain” by Joseph Ecker, Ph.D. of Salk Institute gave a talk on differential expression in adult vs. fetal brain development at Future of Genomics VI Medicine on March 7, 2013.
I like to give snapshot of his talk, and relating to the third wheel of the epigenetics: non coding RNAs for epigenetics, stem cell biology and development. He also reconnecting the dots and demonstrated that there is a linear relation between gene regulation region and methylation type. As a result the plasticity of development takes place with the extensive mutation reconfigurations during early post natal stages up to two years at synaptogenesis.
Ecker’s Study
The study focused onto inheritability of methylation in different organisms and comparative expression pattern. The data from chip sequencing for
- histone modifications,
- whole genome bisulfide sequencing for DNA modifications and
- methylome profiling
projected a differential expression pattern between
for 5 hydroxymethyl cytosine hmC and 5 methyl cytosine mC.
Completion of base resolution of human methylation and aberrant epigenomic reprogramming in induced stem cells showed that the density of genic mCH is positively correlated with gene expression.
- There was an increased mCH and an elevated gene expression pattern, unlike mCG that the gene is silenced at stem cell differentiation.
- mCH expression was not only tissue specific but also cell specific based on comparative expression study.
- There was no mCH expression in fetal frontal cortex unlike adult frontal cortex with accumulation of mCH during synaptogenesis. Also
- deserts of methylation can be counted as heterochromatic regions and protective transfactors between mCG and mCH methylation.
- In DNMT pattern showed neurons enriched with mCH but glia was depleted. Furthermore, these
- sites are not randomly but occurring with correlation.
Ecker group also published (Lister et al, 2009) the first genome-wide study in a mammalian genome, from both
- human embryonic stem cells and
- fetal fibroblasts, along with
- comparative analysis of messenger RNA and
- small RNA components of the transcriptome, several
- histone modifications, and
- sites of DNA–protein interaction for several key regulatory factors.
Like the related review paper by Spivakov and Fisher pointed out the search for molecular signatures of ‘stemness’ and pluripotency is becoming important for cell therapy. Thus, there is a huge effort on transcription machinery of key genes during early development and understanding of stem cells, but working on the epigenetic profiles and their interaction with transcription machinery is equally important. This poised but activable factors under the stem cell genome may open new doors for diagnostics and therapies. As a result, “restricted” human diversity will open doors for a personalized medicine and delivery mechanisms.
Until human genome was sequenced the expected number of genes was high but only 1% of genome is read producing about 25000 genes. That brings up three modules to be concerned:
1. Use the RNA wisely as the ancestor of transferable genetic material that viruses used, even human has embedded over 90% natural viral in their genome;
2. Apply epigenetics with all three types from a scratch;
3. Inheritance.
RNA is regulating the methylation on genome through transposons and silencing the genes at transcriptional level to create intergenerational or transgenerational reprograming. This makes sense since after the decision is made there are two intentions passing onto next generation and maintaining the decision consistently. If
- in soma by mitosis to daughter cells, or
- in germline by meiosis
the characters are transferred to the next generation. However, we also need to mind after the fact because no one is choosing what they get let alone not being able to choose their parents. Choosing the healthy tolerance levels in genome for future medicine is the key.
REFERENCES
Borge, F. and Martiense, R.A. “Establishing epigenetic variation during genome reprogramming” RNA Biology, Volume 10, Issue 4, April 2013, doi: org/10.4161/rna.24085
Dalakouras,A. and Wassenegger, M. “Revisiting RNA-directed DNA methylation” RNA Biology, Volume 10, Issue 3 March 2013 Pages 453 – 455 http://dx.doi.org/10.4161/rna.23542
Piferrer, F. “Epigenetics of sex determination and gonadogenesis” Developmental Dynamics 8 FEB 2013 DOI: 10.1002/dvdy.23924
Yang SY, Baxter EM, Van Doren M. “Phf7 controls male sex determination in the Drosophila germline” Dev Cell. 2012 May 15;22(5):1041-51. doi: 10.1016/j.devcel.2012.04.013.
Yongkyu Park, Mitzi I. Kuroda “Epigenetic Aspects of X-Chromosome Dosage Compensation” Science 10 August 2001: Vol. 293 no. 5532 pp. 1083-1085 DOI: 10.1126/science.1063073
Okano M, Xie S, Li E (July 1998). “Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases” Nat Genet 19 (3): 219–20. doi:10.1038/890
Di Croce L, Raker VA, Corsaro M, et al. (2002). “Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor” Science 295 (5557): 1079–82. doi:10.1126/science.1065173
Jun Kawai1,+, Shinji Hirotsune2,3,Kenji Hirose1,3,+,Shinji Fushiki4, Sachihiko Watanabe1,+ and Yoshihide Hayashizaki2,3, “Methylation profiles of genomic DNA of mouse developmental brain detected by restriction landmark genomic scanning (RLGS) method” Nucl. Acids Res. (1993) 21 (24): 5604-5608. doi: 10.1093/nar/21.24.5604
Cui-Jun Zhang, Jin-Xing Zhou, Jun Liu, Ze-Yang Ma, Su-Wei Zhang, Kun Dou, Huan-Wei Huang, Tao Cai, Renyi Liu, Jian-Kang Zhu and Xin-Jian He. “The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis” The EMBO Journal , (22 March 2013) | doi:10.1038/emboj.2013.49
Ryan Lister1,9, Mattia Pelizzola1,9, Robert H. Dowen1, R. David Hawkins2, Gary Hon2, Julian Tonti-Filippini4, Joseph R. Nery1, Leonard Lee2, Zhen Ye2, Que-Minh Ngo2, Lee Edsall2, Jessica Antosiewicz-Bourget5,6, Ron Stewart5,6, Victor Ruotti5,6, A. Harvey Millar4, James A. Thomson5,6,7,8, Bing Ren2,3 & Joseph R. Ecker1 “Differential methylation between stem cells and adult stem cells” Nature 462, 315-322 (19 November 2009) | doi:10.1038/nature08514
Mikhail Spivakov & Amanda G. Fisher “Epigenetic signatures of stem-cell identity” Nature Reviews Genetics 8, 263-271 (April 2007) | doi:10.1038/nrg2046
Louise C Laurent1,2,3,15, Caroline M Nievergelt4,15, Candace Lynch2,3, Eyitayo Fakunle2,3, Julie V Harness5, Uli Schmidt6, Vasiliy Galat7,8, Andrew L Laslett9,10,11, Timo Otonkoski12,13, Hans S Keirstead5, Andrew Schork4, Hyun-Sook Park14 & Jeanne F Loring2 “Restricted human ethnic diversity in human stem cell lines.” Nature Methods 7, 6 – 7 (2010) doi:10.1038/nmeth0110-06
Other related articles on this topic were published on this Open Access Online Scientific Journal, including the following:
Prostate Cancer Cells: Histone Deacetylase Inhibitors Induce Epithelial-to-Mesenchymal Transition
SJ Williams, PhD
http://pharmaceuticalintelligence.com/2012/11/30/histone-deacetylase-inhibitors-induce-epithelial-to-mesenchymal-transition-in-prostate-cancer-cells/
How mobile elements in “Junk” DNA promote cancer. Part 1: Transposon-mediated tumorigenesis.
SJ Williams, PhD
http://pharmaceuticalintelligence.com/2012/10/31/how-mobile-elements-in-junk-dna-prote-cacner-part1-transposon-mediated-tumorigenesis/
Diagnosing Diseases & Gene Therapy: Precision Genome Editing and Cost-effective microRNA Profiling
Aviva Lev-Ari, PhD, RN, March 28, 2013
http://pharmaceuticalintelligence.com/2013/03/28/diagnosing-diseases-gene-therapy-precision-genome-editing-and-cost-effective-microrna-profiling/
Genomics-based cure for diabetes on-the-way
Ritu Saxena, PhD, March 4, 2013
http://pharmaceuticalintelligence.com/2013/03/04/genomics-based-cure-for-diabetes-on-the-way/
How Genes Function
Larry H Bernstein, MD, FACP, March 4, 2013
http://pharmaceuticalintelligence.com/2013/03/04/how-genes-function/
Long noncoding RNA: UCSF Researchers have Uncovered its role in Brain Development and in Neurological Diseases
Aviva Lev-Ari, PhD, RN, April 17, 2013
http://pharmaceuticalintelligence.com/2013/04/17/long-noncoding-rna-ucsf-researchers-have-uncovered-its-role-in-brain-development-and-in-neurological-diseases/
Bibliographies on Genomics by Subject Matter
Genomics and Genetics Articles on this Open Access Online Scientific Journal 2/2012 — 1/2013
Aviva Lev-Ari, PhD, RN, 2/25/2013
http://pharmaceuticalintelligence.com/biomed-e-books/genomics-orientations-for-personalized-medicine/bibliographies-on-genomics/
The Initiation and Growth of Molecular Biology and Genomics – Part I
Larry H Bernstein, MD, FACP, 2/8/2013
http://pharmaceuticalintelligence.com/2013/02/08/the-initiation-and-growth-of-molecular-biology-and-genomics/
CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease – Part IIC
Larry H Bernstein, MD, FACP, February 14, 2013
http://pharmaceuticalintelligence.com/2013/02/14/cracking-the-code-of-human-life-recent-advances-in-genomic-analysis-and-disease/
Genomic Endocrinology and its Future
Sudipta Saha, December 27, 2012
http://pharmaceuticalintelligence.com/2012/12/27/genomic-endocrinology-and-its-future-2/
Exome sequencing of serous endometrial tumors shows recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes
Sudipta Saha, PhD, December 18, 2012
http://pharmaceuticalintelligence.com/2012/12/18/exome-sequencing-of-serous-endometrial-tumors-shows-recurrent-somatic-mutations-in-chromatin-remodeling-and-ubiquitin-ligase-complex-genes
Pancreatic Cancer: Genetics, Genomics and Immunotherapy
Tilda Barlyia, PhD, April 11, 2013
http://pharmaceuticalintelligence.com/2013/04/11/update-on-pancreatic-cancer/
Exome sequencing of serous endometrial tumors shows recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes
Sudipta Saha, December 18, 2012
http://pharmaceuticalintelligence.com/2012/12/18/exome-sequencing-of-serous-endometrial-tumors-shows-recurrent-somatic-mutations-in-chromatin-remodeling-and-ubiquitin-ligase-complex-genes/
Genomics & Genetics of Cardiovascular Disease Diagnoses: A Literature Survey of AHA’s Circulation Cardiovascular Genetics
3/2010 – 3/2013
Aviva Lev-Ari, PhD, RN and Larry H Bernstein, MD, FACP, March 7, 2013
http://pharmaceuticalintelligence.com/2013/03/07/genomics-genetics-of-cardiovascular-disease-diagnoses-a-literature-survey-of-ahas-circulation-cardiovascular-genetics-32010-32013/
What is the Future for Genomics in Clinical Medicine?
Larry H Bernstein, MD, FACP, February 17, 2013
http://pharmaceuticalintelligence.com/2013/02/17/what-is-the-future-for-genomics-in-clinical-medicine/
CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease – Part IIC
Larry H Bernstein, MD, FACP, February 14, 2013
http://pharmaceuticalintelligence.com/2013/02/14/cracking-the-code-of-human-life-recent-advances-in-genomic-analysis-and-disease/
00
Like this:
Like Loading...
Read Full Post »