CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease – Part IIC
Author: Larry H. Bernstein, MD, FCAP, Triplex Medical Science
Part I: The Initiation and Growth of Molecular Biology and Genomics – Part I From Molecular Biology to Translational Medicine: How Far Have We Come, and Where Does It Lead Us?
http://pharmaceuticalintelligence.com/wp-admin/post.php?post=8634&action=edit&message=1
Part II: CRACKING THE CODE OF HUMAN LIFE is divided into a three part series.
Part IIA. “CRACKING THE CODE OF HUMAN LIFE: Milestones along the Way” reviews the Human Genome Project and the decade beyond.
Part IIB. “CRACKING THE CODE OF HUMAN LIFE: The Birth of BioInformatics & Computational Genomics” lays the manifold multivariate systems analytical tools that has moved the science forward to a groung that ensures clinical application.
Part IIC. “CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease “ will extend the discussion to advances in the management of patients as well as providing a roadmap for pharmaceutical drug targeting.
To be followed by:
Part III will conclude with Ubiquitin, it’s role in Signaling and Regulatory Control.
Part IIC of series on CODE OF HUMAN LIFE
CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease
This final paper of Part II concludes a thorough review of the scientific events leading to the discovery of the human genome, the purification and identification of the components of the chromosome and the DNA structure and role in regulation of embryogenesis, and potential targets for cancer.
The first two articles, Part IIA, Part IIB, go into some depth to elucidate the problems and breakthoughs encountered in the Human Genome Project, and the construction of a 3-D model necessary to explain interactions at a distance.
Part IIC, the final article, is entirely concerned with clinical application of this treasure trove of knowledge to resolving diseases of epigenetic nature in the young and the old, chronic inflammatory diseases, autoimmune diseases, infectious disease, gastrointestinal disorders, neurological and neurodegenerative diseases, and cancer.
CRACKING THE CODE OF HUMAN LIFE: Recent Advances in Genomic Analysis and Disease – Part IIC
1. Gene Links to Heart Disease
Recently, large studies have identified some of the genetic basis for important common diseases such as heart disease and diabetes, but most of the genetic contribution to them remains undiscovered. Now researchers at the University of Massachusetts Amherst led by biostatistician Andrea Foulkes have applied sophisticated statistical tools to existing large databases to reveal substantial new information about genes that cause such conditions as high cholesterol linked to heart disease.
Foulkes says, “This new approach to data analysis provides opportunities for developing new treatments.” It also advances approaches
- to identifying people at greatest risk for heart disease. Another important point is that our method is straightforward to use with freely
- available computer software and can be applied broadly to advance genetic knowledge of many diseases.
The new analytical approach she developed with cardiologist Dr. Muredach Reilly at the University of Pennsylvania and others is called “Mixed modeling of Meta-Analysis P-values” or MixMAP. Because it makes use of existing public databases, the powerful new method
- represents a low-cost tool for investigators.
- MixMAP draws on a principled statistical modeling framework and the vast array of summary data now available from genetic association
- studies to formally test at a new, locus-level, association.
While that traditional statistical method looks for one unusual “needle in a haystack” as a possible disease signal, Foulkes and colleagues’
- new method uses knowledge of DNA regions in the genome that are likely to
- contain several genetic signals for disease variation clumped together in one region.
- Thus, it is able to detect groups of unusual variants rather than just single SNPs, offering a way to “call out” gene
- regions that have a consistent signal above normal variation.
http://Science.com/Science News/Identify Genes Linked to Heart Disease/
2. Apolipoprotein(a) Genetic Sequence Variants
The LPA gene codes for apolipoprotein(a), which, when linked with low-density lipoprotein particles, forms lipoprotein(a) [Lp(a)] —
- a well-studied molecule associated with coronary artery disease (CAD). The Lp(a) molecule has both atherogenic and thrombogenic effects in vitro , but the extent to which these translate to differences in how atherothrombotic disease presents is unknown.
LPA contains many single-nucleotide polymorphisms, and 2 have been identified by previous groups as being strongly associated with
- levels of Lp(a) and, as a consequence, strongly associated with CAD.
However, because atherosclerosis is thought to be a systemic disease, it is unclear to what extent Lp(a) leads to atherosclerosis in other arterial beds (eg, carotid, abdominal aorta, and lower extremity),
- as well as to other thrombotic disorders (eg, ischemic/cardioembolic stroke and venous thromboembolism).
Such distinctions are important, because therapies that might lower Lp(a) could potentially reduce forms of atherosclerosis beyond the coronary tree.
To answer this question, Helgadottir and colleagues compiled clinical and genetic data on the LPA gene from thousands of previous
- participants in genetic research studies from across the world. They did not have access to Lp(a) levels, but by knowing the genotypes for
- 2 LPA variants, they inferred the levels of Lp(a) on the basis of prior associations between these variants and Lp(a) levels. [1]
Their studies included not only individuals of white European descent but also a significant proportion of black persons, in order to
- widen the generalizability of their results.
Their main findings are that LPA variants (and, by proxy, Lp(a) levels) are associated with
- CAD,
- peripheral arterial disease,
- abdominal aortic aneurysm,
- number of CAD vessels,
- age at onset of CAD diagnosis, and
- large-artery atherosclerosis-type stroke.
They did not find an association with
- cardioembolic or small-vessel disease-type stroke;
- intracranial aneurysm;
- venous thrombosis;
- carotid intima thickness; or,
- in a small subset of individuals, myocardial infarction.
Apolipoprotein(a) Genetic Sequence Variants Associated With Systemic Atherosclerosis and Coronary Atherosclerotic Burden but Not With Venous Thromboembolism. Helgadottir A, Gretarsdottir S, Thorleifsson G, et al. J Am Coll Cardiol. 2012;60:722-729

English: Structure of the LPA protein. Based on PyMOL rendering of PDB 1i71. (Photo credit: Wikipedia)
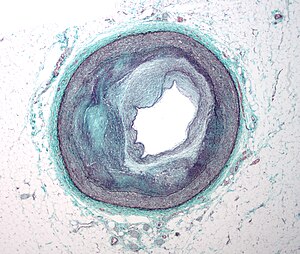
Micrograph of an artery that supplies the heart with significant atherosclerosis and marked luminal narrowing. Tissue has been stained using Masson’s trichrome. (Photo credit: Wikipedia)
Genomic Blueprint of the Heart
Scientists at the Gladstone Institutes have revealed the precise order and timing of hundreds of genetic “switches” required to construct a fully
- functional heart from embryonic heart cells — providing new clues into the genetic basis for some forms of congenital heart disease.
In a study being published online today in the journal Cell, researchers in the laboratory of Gladstone Senior Investigator Benoit Bruneau, PhD,
- employed stem cell technology, next-generation DNA sequencing and computing tools to piece together the instruction manual, or “genomic
- blueprint” for how a heart becomes a heart. These findings offer renewed hope for combating life-threatening heart defects such as arrhythmias (irregular heart beat) and ventricular septal defects (“holes in the heart”).
ScienceDaily (Sep. 13, 2012)
They approach heart formation with a wide-angle lens by
- looking at the entirety of the genetic material that gives heart cells their unique identity.
The news comes at a time of emerging importance for the biological process called “epigenetics,” in which a non-genetic factor impacts a cell’s genetic
- makeup early during development — but sometimes with longer-term consequences. All of the cells in an organism contain the same DNA, but the
- epigenetic instructions encoded in specific DNA sequences give the cell its identity. Epigenetics is of particular interest in heart formation, as the
- incorrect on-and-off switching of genes during fetal development can lead to congenital heart disease — some forms of which may not be apparent until adulthood.
the scientists took embryonic stem cells from mice and reprogrammed them into beating heart cells by mimicking embryonic development in a petri dish. Next, they extracted the DNA from developing and mature heart cells, using an advanced gene-sequencing technique called ChIP-seq that lets scientists “see” the epigenetic signatures written in the DNA.
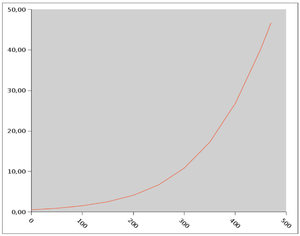
Estimated propability of death or non-fatal myocardial-infarction over one year corresponding ti selectet values of the individual scores. Ordinate: individual score, abscissa: Propability of death or non-fatal myocardial infarction in 1 year (in %) (Photo credit: Wikipedia)
simply finding these signatures was only half the battle — we next had to decipher which aspects of heart formation they encoded
To do that, we harnessed the computing power of the Gladstone Bioinformatics Core. This allowed us to take the mountains of data collected from
- gene sequencing and organize it into a readable, meaningful blueprint for how a heart becomes a heart.”
http://ScienceDaily.org/Scientists Map the Genomic Blueprint of the Heart. ScienceDaily.
Performance of transcription factor identification tools from differential gene expression data
A three step process is a clear way to establish belief in the performance of transcription factor identification tools
- from differential gene expression data.
- identify several types of differential gene expression data sets where the stimulus or trigger is clearly know
- identify the transcription factors most likely associated with the sets expression data.
- perform an upstream analysis from the identified transcription factor.
If the transcription factor and upstream analysis tools can trace the signal cascade back to the stimulus, the tools are
- clearly producing relevant results, and belief in the performance of the analysis tools is established.
At this point, the tools can be directed with confidence to more challenging analyses such as
- developed resistance or pathway elucidation.
The performance of IPA‘s new Transcription Factor and Upstream analysis tools was evaluated on the following datasets (processing details below):
- TGFb stimulation, 1 hour, A549 lung adenocarcinoma cell line
- BMP2 stimulation, 1 hour, Mouse Embryonic Stem Cell E14Tg2A.4
- TNFa stimulation, 1 hour primary murine hepatocytes
For each of the above datasets, an upstream analysis from the identified transcription factors correctly identified the stimulus. IPA’s tools were very
- easy to use and the
- analysis time for the above experiments was less than one minute.
The performance, speed, and ease of use can only be characterized as very good, perhaps leading to breakthroughs when extended and used creatively. Ingenuity’s new transcription factor analysis tool in IPA, coupled with Ingenuity’s established upstream grow tools, should be strongly considered for every lab analyzing differential expression data.
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17896
http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE2639
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19272
Differential expression data was obtained from CEL files using the Matlab functions:
affyrma, genelowvalfilter, genevarfilter, mattest, and mavolcanoplot.
Rick Stanton, Pathway Analysis Consultant Ingenuity.com
3. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells.
Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. J Biol Chem. 2011 Nov 25;286(47):40725-33. Epub 2011 Sep 16.
http://JBiolChem.com/miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells.
NF-E2-related factor 2 (Nrf2) is an important transcription factor that
- activates the expression of cellular detoxifying enzymes.
Nrf2 expression is largely regulated through the association of Nrf2 with Kelch-like ECH-associated protein 1 (Keap1), which
- results in cytoplasmic Nrf2 degradation.
Conversely, little is known concerning the regulation of Keap1 expression. Until now, a regulatory role for microRNAs (miRs) in controlling Keap1 gene expression had not been characterized. By using miR array-
- based screening, we observed miR-200a silencing in breast cancer cells and
- demonstrated that upon re-expression, miR-200a
- targets the Keap1 3′-untranslated region (3′-UTR), leading to Keap1 mRNA degradation. Loss of this regulatory mechanism may
- contribute to the dysregulation of Nrf2 activity in breast cancer. Previously, we have identified epigenetic repression of miR-200a
in breast cancer cells. Here, we find that treatment with epigenetic therapy, the histone deacetylase inhibitor suberoylanilide hydroxamic acid, restored miR-200a expression and reduced Keap1 levels. This reduction in Keap1 levels corresponded with
- Nrf2 nuclear translocation
- and activation of Nrf2-dependent NAD(P)H-quinone oxidoreductase 1 (NQO1) gene transcription.
Moreover, we found that Nrf2 activation inhibited the anchorage-independent growth of breast cancer cells. Finally, our in vitro observations were confirmed in a model of carcinogen-induced mammary hyperplasia in vivo. In conclusion, our study demonstrates
- that miR-200a regulates the Keap1/Nrf2 pathway in mammary epithelium, and we find that epigenetic therapy can restore miR-200a
- regulation of Keap1 expression,
- reactivating the Nrf2-dependent antioxidant pathway in breast cancer.
Nuclear factor-like 2 (erythroid-derived 2, also known as NFE2L2 or Nrf2, is a transcription factor that in humans is encoded by the NFE2L2 gene.[1]) NFE2L2 induces the expression of various genes including those that encode for several antioxidant enzymes, and it may play a physiological role in the regulation of oxidative stress. Investigational drugs that target NFE2L2 are of interest as potential therapeutic interventions for
- oxidative-stress related pathologies.
4. Highly active zinc finger nucleases by extended modular assembly
MS Bhakta, IM Henry, DG Ousterout, KT Das, et al. Corresponding author; email: djsegal@ucdavis.edu
http://CSHNLpress.com/Highly active zinc finger nucleases by extended modular assembly
Zinc finger nucleases (ZFNs) are important tools for genome engineering. Despite intense interest by many academic groups,
- the lack of robust non-commercial methods has hindered their widespread use. The modular assembly (MA) of ZFNs from
- publicly-available one-finger archives provides a rapid method to create proteins that can recognize a very broad spectrum of DNA sequences.
However, three- and four-finger arrays often fail to produce active nucleases. Efforts to improve the specificity of the one-finger archives have not increased the success rate above 25%, suggesting that the MA method might
- be inherently inefficient due to its insensitivity to context-dependent effects.
Here we present the first systematic study on the effect of array length on ZFN activity. ZFNs composed of six-finger MA arrays produced mutations at 15 of 21 (71%) targeted
- loci in human and mouse cells. A novel Drop-Out Linker scheme was used to rapidly assess three- to six-finger combinations,
- demonstrating that shorter arrays could improve activity in some cases. Analysis of 268 array variants revealed that half of
MA ZFNs of any array composition that exceed an ab initio
- B-score cut-off of 15 were active.
- MA ZFNs are able to target more DNA sequences with higher success rates than other methods.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date http://genome.cshlp.org/site/misc/terms.xhtml
After six months, it is available under a Creative Commons License (Attribution-NonCommercial 3.0 Unported License), as described at
http://creativecommons.org/licenses/by-nc/3.0/Highly_active_zinc_finger_nucleases_by_extended_ modular_assembly/
PERSONALIZED MEDICINE in the Pipeline
These insightful reviews are based on the strategic data and insights from Thomson Reuters Cortellis™ for Competitive Intelligence. (A Review of April-June 2012).
The majority of diseases are complex and multi-factorial, involving multiple genes interacting with environmental factors. At the genetic level,
- information from genome-wide association studies that elucidate common patterns of genetic variation across various human populations,
- in addition to profiling, technologies can be utilized in discovery research to provide snapshots of genes and expression profiles that are controlled
- by the same regulatory mechanism and are altered between healthy and diseased states.
The characterization of genes that are abnormally expressed in disease tissues could further be employed as
- diagnostic markers,
- prognostic indicators of efficacy and/or toxicity, or as
- targets for therapeutic intervention.
As the defining catalyst that exponentially paved the way for personalized medicine, information from the published genome sequence revealed that much of the genetic variations in humans are concentrated in about 0.1 percent of the over 3 billion base pairs in the haploid DNA. Most of these variations involve substitution of a single nucleotide for another at a given location in the genetic sequence, known as single nucleotide polymorphism (SNP).
- Combinations of linked SNPs aggregate together to form haplotypes and
- together these serve as markers for locating genetic variations in DNA sequences.
SNPs located within the protein-coding region of a gene or within the control regions of DNA that regulate a gene’s activity could
- have a substantial effect on the encoded protein and thus influence phenotypic outcomes.
Analyzing SNPs between patient population cohorts could highlight specific genotypic variations which can be correlated with specific phenotypic variations in disease predisposition and drug responses.
Prior to the genomic revolution, many of the established therapies were directed against less than 500 drug targets, with many of the top selling drugs acting on well defined protein pathways. However, the sequencing of the human genome has massively expanded the pool of molecular targets that could be exploited in unmet medical needs and currently, of the approximately 22,300 protein-coding genes in the human code, it has been estimated that up to 3000 are druggable. Furthermore, genomic technologies such as
- high-throughput sequencing
- and transcription profiling,
can be used to identify and validate biologically relevant target molecules, or can be applied to cell-based and mice disease models or directly to in vivo human tissues,
- helping to correlate gene targets with phenotypic traits of complex diseases.
This is particularly important, as
- insufficient validation of target gene/proteins in complex diseases may be a contributing factor in the decline in R&D productivity.
Personalized medicine no doubt is already having a tremendous impact on drug development pipelines. According to a study conducted by the Tufts Center for the Study of Drug Development, more than 90 percent of biopharmaceutical companies now utilize at least some
- genomics-derived targets in their drug discovery programs.
However, pipeline analysis from Cortellis for Competitive Intelligence suggests that there is still a scientific gap that has resulted in difficulty optimizing these novel genomic targets into the clinical R&D portfolios of major pharmaceutical companies, particularly outside the oncology field. Selected examples of personalized medicine product candidates in clinical development include (see TABLE 4).
Table 4: Selected Personalized Medicines in Clinical Development
(DATA are Derived from Cortellis for Competitive Intelligence & Thomson Reuters IntegritySM)
http://Thomson Reuters.com/Cortellis for Competitive Intelligence/IntegritySM/Table_4_Selected_Personalized_Medicines_in_Clinical_Development/
PHARMA MATTERS | SPOTLIGHT ON… PERSONALIZED MEDICINE
The paucity of actual targeted therapy examples, especially outside oncology, suggest
- that integration of the personalized medicine paradigm into biopharmaceutical R&D is still fraught with challenges.
Despite the fact that the Human genome Project has been completed for over ten years, the broader application of genomics with drug development
- still remains unrealized, and is hampered by a number of scientific challenges. One of the major obstacles stems from
- incomplete association of genomic alterations with complex disease pathways and the phenotypic consequences.
As the modality of most complex diseases are multi-factorial, understanding how each genomic driver event plays a role in disease and the
- interaction/interdependence with other genetic and environmental factors is important for
- determining the rationale for targeted prevention or treatment of the disease.
Mutations found in Melanomas may shed light on Cancer Growth
Gina Kolata. New York Times.
http://NewYorkTimes.com/mutations_found_in_melanomas_may_shed-light_on_how_cancers_grow/
Mutations in Melanoma are in regions that control genes, not in the genes themselves. The mutations are exactly the type caused by exposure to ultraviolet light. The findings are reported in two papers in http://Science.com/ScienceExpress/
A German Team led by Rajiv Kumar (Heidelberg) and Dirk Schadendorf (Essen) looked at a family whose members tended to get melanomas. Their findings indicate that those inherited with the mutations might be born with cells that have taken the first step toward cancer.
Comment
This discussion addresses the issues raised about the direction to follow in personalized medicine. Despite the amount of work necessary to bring the clarity that is sought after, the experiments and experimental design is most essential.
- The arrest of ciliogenesis in ovarian cancer cell lines compared to wild type (WT) ovarian epithelial cells, and
- The link to suppressing ciliogenesis by AURA protein and CHFR at the base of the cilium, which disappears at mitosis or with proliferation.
- There is no accumulation by upregulation of PDGF under starvation by the cancer cells compared to the effect in WT OSE.
Here we have a systematic combination of signaling events tied to changes in putative biomarkers that occur synchronously in Ov cancer cell lines.
These changes are identified with changes in
- proliferation,
- loss of ciliary structure, and
- proliferation.
In this described scenario,
- WT OSE cells would be arrested, and
- it appears that they would take the path to apoptosis (under starvation).
Even without more information, this cluster is what one wants to have in a “syndromic classification”. The information used to form the classification entails the identification of strong ‘signaling-related’ biomarkers. The Gli2 peptide has to be part of this.
In principle, a syndromic classification would be ideally expected to have no less than 64 classes. If the classification is “weak”, then the class frequencies would be close to what one would expect in the WT OSE. In this case, in reality,
- several combinatorial classes would have low frequency, and
- others would be quite high.
This obeys the classification rules established by feature identification, and the information gain described by Solomon Kullback and extended by Akaike.
Does this have to be the case for all different cancer types? I don’t think so. The cells are different in ontogenesis. In this case, even the WT OSE have mesenchymal features and so, are not fully directed to epithelial expression. This happens to be the case in actual anatomic expression of the ovary. On the other hand, one would expect shared features of the
- ovary,
- testes,
- thyroid,
- adrenals, and
- pituitary.
There is biochemical expression in terms of their synthetic function – TPN organs. I would have to put the liver into that broad class. Other organs – skeletal muscle & heart – transform substrate into energy or work. (Where you might also put intestinal smooth muscle).
They have to have different biomarker expressions, even though they much less often don’t form neoplasms. (Bone is not just a bioenergetic force. It is maintained by muscle action. It forms sarcomas. But there has to be a balance between bone removal by osteoclasts and refill by osteoblasts.)
Viewpoint: What we have learned
- The Watson-Crick model proposed in 1953 is limited for explaining fully genome effects
- The Pauling triplex model may have been prescient because of a more full anticipation of molecular bonding variants
- A more adequate triple-helix model has been proposed and is consistent with a compact genome in the nucleus
The structure of the genome is not as we assumed – based on the application of Fractal Geometry. Current body of evidence is building that can reveal a more complete view of genome function.
- transcription
- cell regulation
- mutations
Summary
I have just completed a most comprehensive review of the Human Genome Project. There are key research collaborations, problems in deciphering the underlying structure of the genome, and there are also both obstacles and insights to elucidating the complexity of the final model.
This is because of frequent observations of molecular problems in folding and other interactions between nucleotides that challenge the sufficiency of the original DNA model proposed by Watson and Crick. This has come about because of breakthrough innovation in technology and in computational methods.
Radoslav Bozov •
Molecular biology and growth was primarily initiated on biochemical structural paradigms aiming to define functional spatial dynamics of molecules via assignation of various types of bondings – covalent and non-covalent – hydrogen, ionic , dipole-dipole, hydrophobic interactions.
- Lab techniques based on z/m paradigm allowed separation, isolation and identification of bio substances with a general marker identity finding correlation between physiological/cellular states.
- The development of electronic/x-ray technologies allowed zooming in nano space without capturing time.
- NMR technology identified the existence of space topology of initial and final atomic states giving a highly limited light on time – energy axis of atomic interactions.
- Sequence technology and genomic perturbations shed light on uncertainty of genomic dynamics and regulators of functional ever expanding networks.
- Transition state theory coupled to structural complexity identification and enzymatic mechanisms ran up parallel to work on various phenomena of strings of nucleotides (oligomers and polymers) – illusion/observation of constructing models on the dynamics of protein-dna-rna interference.
- The physical energetic constrains of biochemistry were inapplicable in open biological systems. Biologists have accepted observation as a sole driver towards re-evaluating models.
- The separation of matter and time constrains emerged as deviation of energy and space constrains transforming into the full acceptance of code theory of life. One simple thing was left unnoticed over time –
- the amount of information of quantum matter within a single codon is larger than that of a single amino acid. This violated all physical laws/principles known to work with a limited degree of certainty.
- The limited amount of information analyzed by conventional sequence identity led to the notion of applicability of statistical measures of and PCR technology. Mutations were identified over larger scale of data.
- Quantum chemistry itself is being limited due discrete space/energy constrains, thus it transformed into concepts/principles in biology that possess highly limited physical values whatsoever.
- The central dogma is partially broken as a result of
- regulatory constrains
- epigenetic phenomena and
- iRNA.
Large scale code computational data run into uncertainty of the processes of evolution and its consequence of signaling transformation. All drugs were ‘lucky based’ applicability and/or discovery with largely unpredictable side effect over time.
Other Related articles on this Open Access Online Sceintific Journal include the following:
Big Data in Genomic Medicine lhb
http://pharmaceuticalintelligence.com/2012/12/17/big-data-in-genomic-medicine/
BRCA1 a tumour suppressor in breast and ovarian cancer – functions in transcription, ubiquitination and DNA repair S Saha http://pharmaceuticalintelligence.com/2012/12/04/brca1-a-tumour-suppressor-in-breast-and-ovarian-cancer-functions-in-transcription-ubiquitination-and-dna-repair/
Computational Genomics Center: New Unification of Computational Technologies at Stanford A Lev-Ari http://pharmaceuticalintelligence.com/2012/12/03/computational-genomics-center-new-unification-of-computational-technologies-at-stanford/
Personalized medicine gearing up to tackle cancer ritu saxena http://pharmaceuticalintelligence.com/2013/01/07/personalized-medicine-gearing-up-to-tackle-cancer/
Differentiation Therapy – Epigenetics Tackles Solid Tumors sj Williams http://pharmaceuticalintelligence.com/2013/01/03/differentiation-therapy-epigenetics-tackles-solid-tumors/
Mechanism involved in Breast Cancer Cell Growth: Function in Early Detection & Treatment A Lev-Ari http://pharmaceuticalintelligence.com/2013/01/17/mechanism-involved-in-breast-cancer-cell-growth-function-in-early-detection-treatment/
The Molecular pathology of Breast Cancer Progression tilde barliya http://pharmaceuticalintelligence.com/2013/01/10/the-molecular-pathology-of-breast-cancer-progression/
Gastric Cancer: Whole-genome reconstruction and mutational signatures A Lev-Ari http://pharmaceuticalintelligence.com/2012/12/24/gastric-cancer-whole-genome-reconstruction-and-mutational-signatures-2/
Paradigm Shift in Human Genomics – Predictive Biomarkers and Personalized Medicine – Part 1 (pharmaceuticalintelligence.com) A Lev-Ari http://pharmaceuticalintelligence.com/2013/01/13/paradigm-shift-in-human-genomics-predictive-biomarkers-and-personalized-medicine-part-1/
LEADERS in Genome Sequencing of Genetic Mutations for Therapeutic Drug Selection in Cancer Personalized Treatment: Part 2 A Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/leaders-in-genome-sequencing-of-genetic-mutations-for-therapeutic-drug-selection-in-cancer-personalized-treatment-part-2/
Personalized Medicine: An Institute Profile – Coriell Institute for Medical Research: Part 3 A Lev-Ari http://pharmaceuticalintelligence.com/2013/01/13/personalized-medicine-an-institute-profile-coriell-institute-for-medical-research-part-3/
Harnessing Personalized Medicine for Cancer Management, Prospects of Prevention and Cure: Opinions of Cancer Scientific Leaders @ http://pharmaceuticalintelligence.com ALA http://pharmaceuticalintelligence.com/2013/01/13/7000/Harnessing Personalized Medicine for Cancer Management, Prospects of Prevention and Cure: Opinions of Cancer Scientific Leaders/
GSK for Personalized Medicine using Cancer Drugs needs Alacris systems biology model to determine the in silico effect of the inhibitor in its “virtual clinical trial” A Lev-Ari http://pharmaceuticalintelligence.com/2012/11/14/gsk-for-personalized-medicine-using-cancer-drugs-needs-alacris-systems-biology-model-to-determine-the-in-silico-effect-of-the-inhibitor-in-its-virtual-clinical-trial/
Recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes in serous endometrial tumors S Saha http://pharmaceuticalintelligence.com/2012/11/19/recurrent-somatic-mutations-in-chromatin-remodeling-and-ubiquitin-ligase-complex-genes-in-serous-endometrial-tumors/
Personalized medicine-based cure for cancer might not be far away ritu saxena http://pharmaceuticalintelligence.com/2012/11/20/personalized-medicine-based-cure-for-cancer-might-not-be-far-away/
Human Variome Project: encyclopedic catalog of sequence variants indexed to the human genome sequence A Lev-Ari
http://pharmaceuticalintelligence.com/2012/11/24/human-variome-project-encyclopedic-catalog-of-sequence-variants-indexed-to-the-human-genome-sequence/
Prostate Cancer Cells: Histone Deacetylase Inhibitors Induce Epithelial-to-Mesenchymal Transition sjwilliams
http://pharmaceuticalintelligence.com/2012/11/30/histone-deacetylase-inhibitors-induce-epithelial-to-mesenchymal-transition-in-prostate-cancer-cells/
Inspiration From Dr. Maureen Cronin’s Achievements in Applying Genomic Sequencing to Cancer Diagnostics A Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/10/inspiration-from-dr-maureen-cronins-achievements-in-applying-genomic-sequencing-to-cancer-diagnostics/
The “Cancer establishments” examined by James Watson, co-discoverer of DNA w/Crick, 4/1953 A Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/09/the-cancer-establishments-examined-by-james-watson-co-discover-of-dna-wcrick-41953/
Directions for genomics in personalized medicine lhb http://pharmaceuticalintelligence.com/2013/01/27/directions-for-genomics-in-personalized-medicine/
How mobile elements in “Junk” DNA promote cancer. Part 1: Transposon-mediated tumorigenesis. Sjwilliams
http://pharmaceuticalintelligence.com/2012/10/31/how-mobile-elements-in-junk-dna-prote-cancer-part1-transposon-mediated-tumorigenesis/
Mitochondria: More than just the “powerhouse of the cell” eritu saxena http://pharmaceuticalintelligence.com/2012/07/09/mitochondria-more-than-just-the-powerhouse-of-the-cell/
Mitochondrial fission and fusion: potential therapeutic targets? Ritu saxena http://pharmaceuticalintelligence.com/2012/10/31/mitochondrial-fission-and-fusion-potential-therapeutic-target/
Mitochondrial mutation analysis might be “1-step” away ritu saxena http://pharmaceuticalintelligence.com/2012/08/14/mitochondrial-mutation-analysis-might-be-1-step-away/
mRNA interference with cancer expression lhb http://pharmaceuticalintelligence.com/2012/10/26/mrna-interference-with-cancer-expression/
Related articles
- Researchers Find Gene Variant Linked To Aortic Valve Disease (medicalnewstoday.com)
- xR: The Human Genome Project has Delivered on its Promise to Treat Disease. Introducing GEMS, the Genetic Enzyme Methylation Syndrome (prweb.com)
- Biostatisticians Identify Genes Linked To Heart Disease (medicalnewstoday.com)
- Four barriers that must fall before the personalized medicine revolution can start (medcitynews.com)
- The personalized medicine revolution is almost here (venturebeat.com)


