DNA Structure and Oligonucleotides
Curator: Larry H Bernstein, MD, FCAP
Triplex Medical Science
Expert, Author, Writer, Leaders in Pharmaceutical Business Intelligence
http:/pharmaceuticalintelligence.com/DNA_structure_ and_ Oligonucleotides

A section of DNA; the sequence of the plate-like units (nucleotides) in the center carries information. (Photo credit: Wikipedia)
Triplex DNA
1. A Third Strand for DNA
The DNA double helix can under certain conditions accommodate a third strand in its major groove. Researchers in the UK have now presented a complete set of
- four variant nucleotides that makes it possible to use this phenomenon in gene regulation and mutagenesis.
Natural DNA only forms a triplex if the targeted strand is rich in purines – guanine (G) and adenine (A) – which in addition to the bonds of the Watson-Crick base pairing can form two further hydrogen bonds, and
- the ‘third strand’ oligonucleotide has the matching sequence of pyrimidines – cytosine (C) and thymine (T).
Any Cs or Ts in the target strand of the duplex will only bind very weakly, as they contribute just one hydrogen bond. Moreover, the recognition of G requires the C in the probe strand to be protonated, so triplex formation will only work at low pH.
To overcome all these problems, the groups of Tom Brown and Keith Fox at the University of Southampton have developed modified building blocks, and have now completed a set of
- four new nucleotides, each of which will bind to one DNA nucleotide from the major groove of the double helix.1
They tested the binding of a 19-mer of these designer nucleotides to a double helix target sequence in comparison with the corresponding triplex-forming oligonucleotide made from natural DNA bases. Using fluorescence-monitored thermal melting and DNase I footprinting, the researchers showed that their construct forms stable triplex even at neutral pH. Tests with mutated versions of the target sequence showed that
- three of the novel nucleotides are highly selective for their target base pair,
- while the ‘S’ nucleotide, designed to bind to T, also tolerates C.
In principle, triplex formation has already been demonstrated as a way of inducing mutations in cell cultures and animal experiments.2
Michael Gross
References
1 DA Rusling et al, Nucleic Acids Res. 2005, 33, 3025 http://NucleicAcidsRes.com/Rusling_DA
2 KM Vasquez et al, Science 2000, 290, 530 http://Science.org/Vazquez_KM
2. Triplex DNA Structures
Triplex DNA Structures. Frank-Kamenetskii, Mirkin SM. Annual Rev Biochem 1995; 64:69-95./www.annualreviews.org/aronline
Since the pioneering work of Felsenfeld, Davies, & Rich (1), double-stranded polynucleotides containing
- purines in one strand
- and pydmidines in the other strand [such as poly(A)/poly(U), poly(dA)/poly(dT), or poly(dAG)/poly(dCT)]
have been known to be able to undergo a stoichiometric transition forming a triple-stranded structure containing one polypurine and two polypyrimidine strands. Early on, it was assumed that the third
strand was located in the major groove and associated with the duplex via non-Watson-Crick interactions now known as Hoogsteen pairing.
Triple helices consisting of one pyrimidine and two purine strands were also proposed. However, notwithstanding the fact that single-base triads in tRNAs tructures were well-documented, triple-helical DNA escaped wide attention before the mid-1980s.
The considerable modern interest in DNA triplexes arose due to two partially independent developments.
First, homopurine-homopyrimidine stretches in supercoiled plasmids were found
- to adopt an unusual DNA structure, called
- H-DNA which includes a triplex as the major structural element.
Secondly, several groups demonstrated that homopyrimidine and some
- purine-rich oligonucleotidescan form stable and sequence-specific complexes with
- corresponding homopurine-homopyrimidine sites on duplex DNA. These
complexes were shown to be triplex structures rather than D-loops, where the
- oligonucleotide invades the double helix and displaces one strand.
A characteristic feature of all these triplexes is that the two chemically homologous strands (both pyrimidine or both purine) are antiparallel. These findings led explosive growth in triplex studies.
One can easily imagine numerous “geometrical” ways to form a triplex, and those that have been studied experimentally. The canonical intermolecular triplex consists of either
- three independent oligonucleotide chains or
- of a long DNA duplex carrying homopurine-homopyrimidine insert
- and the corresponding oligonucleotide.
Triplex formation strongly depends on the oligonucleotide(s) concentration. A single DNA chain may also fold into a triplex connected by two loops. To comply with the sequence and
- polarity requirements for triplex formation, such a DNA strand must have a peculiar sequence: It contains
- a mirror repeat (homopyrimidine for YR*Y triplexes and homopurine for YR*R triplexes)
- flanked by a sequence complementary to one half of this repeat.
Such DNA sequences fold into triplex configuration much more readily than do the corresponding intermolecular triplexes, because all triplex forming segments are brought together within the same molecule.
It has become clear recently, however, that both sequence requirements and chain polarity rules for triplex formation can be met by DNA target sequences built
of clusters of purines and pyrimidines. The third strand consists of
- adjacent homopurine and homopyrimidine blocks forming Hoogsteen hydrogen bonds with purines
- on alternate strands of the target duplex, and this strand switch preserves the proper chain polarity.
- These structures, called alternate-strand triplexes, have been
experimentally observed as both intra- and intermolecular triplexes. These results increase the number of potential targets for triplex formation in natural DNAs somewhat by
- adding sequences composed of purine and pyrimidine clusters, although
- arbitrary sequences are still not targetable because strand switching is energetically unfavorable.
REFERENCES
Lyamichev VI, Mirkin SM, Frank-Kamenetskii MD. J. Biomol. Stract. Dyn. 1986; 3:667-69. http://JbiomolStractDyn.com/Lyamichev_VI/
Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN0 Filippov SA, Frank-Kamenetskii MD. Nature 1987; 330:495-97. http://Nature.com/
Demidov V, Frank-Kamenetskii MD, Egholm M, Buchardt O, Nielsen PE. Nucleic Acids Res. 1993; 21:2103-7. http://NucleicAcidsResearch.com/
Mirkin SMo Frank-Kamenetskii MD. Anna. Rev. Biophys. Biomol. Struct. 1994; 23:541-76. http://AnnRevBiophysBiomolecStructure.com/
Hoogsteen K. Acta Crystallogr. 1963; 16:907-16 http://ActaCrystallogr.com/
Malkov VA, Voloshin ON, Veselkov AG, Rostapshov VM, Jansen I, et al. Nucleic Acids Res. 1993; 21:105-11. http://NucleicAcidsResearch.com/
Malkov VA, Voloshin ON, Soyfer VN, Frank-Kamenetskii MD. Nucleic Acids Res. 1993; 21:585-91
Chemy DY, Belotserkovskii BP, Frank-Kamenetskii MD, Egholm M, Buchardt O, et al. Proc. Natl. Acad. Sci. USA 1993; 90:1667-70 http://PNAS.org/
3. Triplex forming Oligonucleotides
Triplex forming oligonucleotides: sequence-specific tools for genetic targeting. Knauert MP, Glazer PM. Human Molec Genetics 2001; 10(20):2243-2251.
Triplex forming oligonucleotides (TFOs) bind in the major groove of duplex DNA with a high specificity and affinity. Because of these characteristics, TFOs have been proposed as homing devices for genetic manipulation in vivo.
These investigators review work demonstrating the ability of TFOs and related molecules to
- alter gene expression and
- mediate gene modification in mammalian cells.
- TFOs can mediate targeted gene knock out in mice,
providing a foundation for potential application of these molecules in human gene therapy.
4. Novagon DNA
John Allen Berger, founder of Novagon DNA and The Triplex Genetic Code
Over the past 12+ years, Novagon DNA has amassed a vast array of empirical findings which challenge the “validity” of the “central dogma theory”, especially the current five nucleotide Watson-Crick DNA and RNA genetic codes. DNA = A1T1G1C1, RNA =A2U1G2C2.
We propose that our new Novagon DNA 6 nucleotide Triplex Genetic Code has more validity than the existing 5 nucleotide (A1T1U1G1C1) Watson-Crick genetic codes. Our goal is to conduct a “world class” validation study to replicate and extend our findings.
Triplex DNA Structures
Maxim D. Frank-Kamenetskii, Sergei M. Mirkin
Perspectives and Summary
Doubling down: four-stranded, ‘quadruple helix’ DNA discovered
Electrochemical Determination of Triple Helices: Electrocatalytic Oxidation of Guanine in an IntramolecularTriplex
Rebecca C. Holmberg and H. Holden Thorp
Triplex DNA: fundamentals, advances, and potential applications for gene therapy
Phillip P. Chan, P. M. Glazer
http://JMolecMed.com/Triplex DNA: fundamentals, advances, and potential applications for gene therapy/
A Gold Nanoparticle Based Approach for Screening Triplex DNA Binders
Min Su Han, Abigail K. R. Lytton-Jean, and Chad A. Mirkin*
Systems Integrated Biomedical Research
Cutting a SWATH through Personalized Medicine
Genetics and Biophysics for Large Volumes of Data
Researchers develop Compilation of Protein Interaction Data
Posted on January 2, 2013 by grathbone
Dual coding in alternative reading frames correlates with intrinsic protein disorder
Erika Kovacs, Peter Tompa, Karoly Liliom, and Lajos Kalmar1
Nucleosides
Functionalized Nucleoside 5′-triphosphates for In Vitro Selection of New Catalytic Ribonucleic Acids
JMatulic-Adamic, AT Daniher, A Karpeisky, P Haeberli, D Sweedler and L Beigelman*
http://BioorgMedChemLet.com/Functionalized Nucleoside 5′-triphosphates for In Vitro Selection of New Catalytic Ribonucleic Acids/
Bioorganic & Medicinal Chemistry Letters 10 (2000) 1299±1302
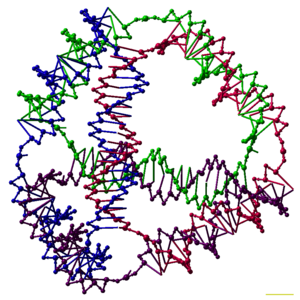
English: A model of a DNA tetrahedron. Each edge of the tetrahedron is a 20bp DNA duplex, and each vertex is a three-arm junction. In this model each basepair is represented by five pseudo-atoms, representing the two sugars, the two phosphates, and the major groove. The scale bar is 1 nm. (Photo credit: Wikipedia)

From left to right, the structures of A-, B- and Z-DNA. The structure a DNA molecule depends on its environment. In aqueous enviromnents, including the majority of DNA in a cell, B-DNA is the most common structure. The A-DNA structure is dominates in dehydrated samples and is similar to the double-stranded RNA and DNA/RNA hybrids. Z-DNA is a rarer structure found in DNA bound to certain proteins. (Photo credit: Wikipedia)
Related articles
- ‘Quadruple helix’ DNA discovered in human cells (sciencedaily.com)
- Scientists successfully store data in DNA (earthsky.org)
- ‘Quadruple helix’ DNA discovered in human cells (phys.org)
- ‘Quadruple helix’ DNA discovered in human cells (esciencenews.com)







I actually consider this amazing blog , âSAME SCIENTIFIC IMPACT: Scientific Publishing –
Open Journals vs. Subscription-based « Pharmaceutical Intelligenceâ, very compelling plus the blog post ended up being a good read.
Many thanks,Annette
I actually consider this amazing blog , âSAME SCIENTIFIC IMPACT: Scientific Publishing –
Open Journals vs. Subscription-based « Pharmaceutical Intelligenceâ, very compelling plus the blog post ended up being a good read.
Many thanks,Annette