Targeted Nucleases
Curator: Larry H Bernstein, MD, FCAP
A REVIEW of 3 published works
Targeted nucleases: spreading the joy
Nature Methods 10, 179 (2013) http://dx.doi.org/10.1038/nmeth.2402
- by a complementary RNA molecule.
- zinc-finger nucleases (ZFNs) and
- transcription activator–like effector nucleases (TALENs),
- target a new sequence.
- the human gene that encodes interferon into a bacterial cell’s genome.
- acquired the human interferon gene proceeded to produce interferon at a rapid rate, and to grow and divide.
The millions of interferon-producing bacteria growing in the culture were all descendants of the cell that had originally received the human interferon gene.
- the ability to cut DNA into recognizable pieces and rearrange those pieces in different ways.
- a piece of DNA carrying the interferon gene was
- inserted into a plasmid,which
- carried the gene into a bacterial cell.
- by enzymes that recognize and
- cleave specific sequences of nucleotides in DNA.
- that fragment the viral DNA as soon as it enters the bacterial cell.
- specific nucleotide sequences in a DNA strand,
- bind to the DNA at those sequences, and
- cleave the DNA at a particular place within the recognition sequence.
- bacteria modify their own DNA, using other enzymes known as methylases to add methyl (CH3) groups
- to some of the nucleotides in the bacterial DNA.
- the endonuclease cannot bind to that sequence.
- the bacterial DNA is protected from being degraded at that site.
- but viral DNA has not been methylated, and therefore
- is not protected from enzymatic cleavage.
- typically four to six nucleotides long, and
- they are often palindromes.
- the nucleotides at one end of the recognition sequence are complementary to those at the other end, so that
- the two strands of the DNA duplex have the same nucleotide sequence running in opposite directions for the length of the recognition sequence.
Two important consequences arise from this arrangement of nucleotides to be discussed.
- theycleave that DNA at defined positions.
- The most well studied class are the so-called type II restriction enzymes.
- First, they must cleave only DNA molecules that contain recognition sites (hereafter referred to as cognate DNA) without cleaving DNA molecules that lack these sites.
- endonucleases must cleave cognate DNA molecules much more than 5000 times as efficiently as they cleave nonspecific sites.
- Second, restriction enzymes must not degrade the host DNA.
How do these enzymes manage to degrade viral DNA while sparing their own?
- These pairs of enzymes are referred to as restriction-modification systems.
- The incoming nucleophile attacks the phosphorus atom, and
- a pentacoordinate transition state is formed.
This species has a trigonal bipyramidal geometry centered at the phosphorus atom, with
- the incoming nucleophile at one apex of the two pyramids and the group that is displaced (the leaving group, L) at the other apex.
- The two mechanisms differ in the number of times the displacement occurs in the course of the reaction.
- a pentacoordinate transition state is formed.
- rules out the formation of any covalently bound intermediate (Figure 9.35).
- bound to oligonucleotides that contain the appropriate recognition sequences.
- when produced, the crystals are soaked in solutions containing the metal.
- No cleavage takes place, allowing the location of the magnesium ion binding sites to be determined (Figure 9.36).
- three are water molecules,
- two are carboxylates of the enzyme’s aspartate residues, and
- one is an oxygen atom of the phosphoryl group at the site of cleavage.
- in conjunction with the aspartate residues,
- helps polarize the water molecule toward deprotonation.
This arrangement gives the three-dimensional structure of the recognition site
- a twofold rotational symmetry (Figure 9.37).
- The matching symmetry of the recognition sequence and the enzyme has been confirmed
- by the determination of the structure of the complex between EcoRV endonuclease and DNA fragments containing its recognition sequence (Figure 9.38).
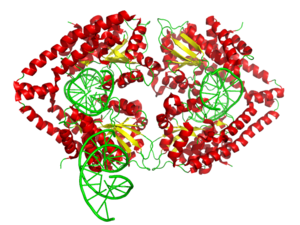
The enzyme surrounds the DNA in a tight embrace.
This view of the structure of EcoRV endonuclease bound to a cognate DNA fragment is down the helical axis of the DNA. The two protein subunits are in yellow and blue, and the DNA backbone is in red.
- by hydrogen bonding with residues that are located in two loops,
- one projecting from the surface of each enzyme subunit (Figure 9.39).
- the enzyme binds to all sequences, both cognate and noncognate, with approximately equal affinity.
- the structures of complexes formed with noncognate DNA fragments are strikingly different from those formed with cognate DNA:
- the noncognate DNA conformation is not substantially distorted (Figure 9.41).
This lack of distortion has important consequences with regard to catalysis. No phosphate is positioned sufficiently close to the active-site aspartate residues to complete a magnesium ion binding site (see Figure 9.36). Hence, the nonspecific complexes do not bind the magnesium ion and
- the complete catalytic apparatus is never assembled.
- the catalytic specificity of more than 1,000,000-fold that is observed for EcoRV endonuclease
- in the nonspecific complex, the DNA backbone is too far from the enzyme
- additional contacts are made between the enzyme and the substrate, increasing the binding energy.
- However, the distortion in the cognate complex dramatically affects catalysis by completing the magnesium ion binding site.
- This example illustrates how enzymes can utilize available binding energy to deform substrates and poise them for chemical transformation.
- Interactions that take place within the distorted substrate complex
- stabilize the transition state leading to DNA hydrolysis.
- the methyl group’s presence precludes the formation of a hydrogen bond between the amino group and the side-chain carbonyl group of asparagine 185 (Figure 9.43).
- This asparagine residue is closely linked to the other amino acids that form specific contacts with the DNA.
- The absence of the hydrogen bond disrupts other interactions between the enzyme and the DNA substrate, and
- the distortion necessary for cleavage will not take place.
- between EcoRV endonuclease and cognate DNA molecules and
- prevents their hydrolysis.
- the presence of a core structure conserved in the different enzymes.
- This structure includes β strands that contain the aspartate (or, in some cases, glutamate) residues forming the magnesium ion binding sites (Figure 9.44).
- from other species by horizontal gene transfer, the passing between species of pieces of DNA (such as plasmids) that provide
- a selective advantage in a particular environment.
- indicative of a close evolutionary relationship.
- these species of bacteria are not closely related,
- as is known from sequence comparisons of other genes and other evidence.
- more recently than the time of their evolutionary divergence.
- the gene encoding EcoRI endonuclease uses particular codons to specify given amino acids that are
- strikingly different from the codons used by most E. coli genes, which
- suggests that the gene did not originate in E. coli.
- Horizontal gene transfer may be a relatively common event.
- genes that inactivate antibiotics are often transferred, leading to the transmission of antibiotic resistance from one species to another.
- protection against viral infections may have favored horizontal gene transfer.
- Cleavage Is by In-Line Displacement of 3′ Oxygen from Phosphorus by Magnesium-Activated Water
- Restriction Enzymes Require Magnesium for Catalytic Activity
- The Complete Catalytic Apparatus Is Assembled Only Within Complexes of Cognate DNA Molecules, Ensuring Specificity
- Type II Restriction Enzymes Have a Catalytic Core in Common and Are Probably Related by Horizontal Gene Transfer

English: 3d surface model of HindIII dimer complexed with a DNA fragment from PDB 2E52. Ref.: Watanabe, N., Sato, C., Takasaki, Y., Tanaka, I. Crystal structural analysis of HindIII restriction endonuclease in complex with cognate DNA at 2.0 angstrom resolution to be published (Photo credit: Wikipedia)
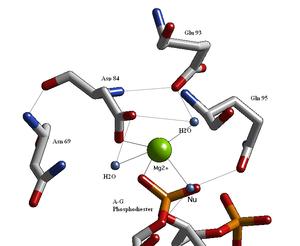
English: BglII active site containing residues that coordinate to a metal ion and water molecules including the nucleophilic water that breaks the scissile phosphodiester bond at the recognition site. (Photo credit: Wikipedia)
Related articles
- Efficient Methods for Targeted Mutagenesis in Zebrafish Using Zinc-Finger Nucleases: Data from Targeting of Nine Genes Using CompoZr or CoDA ZFNs (plosone.org)
- The Promise of Synthetic Biology (dreamerbiologist.wordpress.com)
- Virus Stole Bacteria Immune System Now Kills Resistant Cholera (medicaldaily.com)
- MIT researchers crack cheap, precise gene therapy (extremetech.com)
- New method allows scientists to insert multiple genes in specific locations and delete defective genes (nextbigfuture.com)
- Cheap and easy technique to snip DNA could revolutionize gene therapy (esciencenews.com)
- Monsters, Inc.? New software lets scientists program life (foxnews.com)
- The Virus That Learns (phenomena.nationalgeographic.com)
- Major advance in genetic research (hhmi.org)