Directions for Genomics in Personalized Medicine
Author: Larry H. Bernstein, MD, FCAP

Word Cloud by Daniel Menzin
Purpose
This discussion will identify the huge expansion of genomic technology in the search for biopharmacotherapeutic targets that continue to be explored involving different levels and interacting signaling pathways. There are several methods of analyzing gene expression that will be discussed. Great primary emphasis required investigation of combinations of mutations expressed in different cancer types. James Watson has proposed a major hypothesis that expresses the need to focus on “central” “driver mutations” that correspond with the regulation of gene expression, cell proliferation, and cell metabolism eith a critical rejection of antioxiant benefits. What hasn’t been know is why drug resistance develops and whether the cellular migration and aerobic glycolysis can be redirected after cell metastasis occurs. I attempt to bring out the complexities of current efforts.
.Introduction
- This discussion is a continuation of a previous discussion on the role of genomics if discovery of therapeutic targets for cancer, each somewhat different, but all related to:
- The reversal of carcinoma by targeting a key driver of multiple signaling pathways that activate cell proliferation
- Pinpointing a stage in a multistage process at which tumor progression links to changes in morphology from basal cells to invasive carcinoma with changes in polarity and loss of glandular architecture
- Reversal of the carcinoma through using a small molecule that either is covalently bound to a nanoparticle delivery system that blocks or reverses tumor development
- Synthesis of a small molecule that interacts with the translation of the genome either by substitution of a key driver molecule or by blocking at the mRNA stage of translation
- Blocking more than one signaling pathway that are links to carcinogenesis and cellular proliferation and invasion
Difficulty of the problem
A problem expressed by James Watson is that the investigations that are ongoing
- are following a pathway that is not driven by attacking the “primary” driver of carcinogenesis.
He uses the Myc gene as an example, as noted in the previous discussion. The problem may be more complicated than he envisions.
- The most consistent problem in chemotherapy, irrespective of the design and the target has been cancer remission for a short time followed by recurrence, and then
- switching to another drug, or combination chemotherapy.
It is common to “clean” the field at the time of resection using radiotherapy before chemotherapy.
- But the goal is understood to be “palliation”, not cure.
This raises a serious issue in the hypothesis posed by Watson. The issue is
- whether there is a core locus of genetic regulation that is common to carcinogenesis irrespective of tissue metabolic expression.
- This is supported by the observation that tissue specific express is lost in cancer cells by de-differentiation.
Historical Perspective
AEROBIC GLYCOLYSIS
In 1967 Otto Warburg published his view in a paper “The prime cause and prevention of cancer”.
There are primary and secondary causes of all diseases
- plague – primary: plague bacillus
- plague – secondary: filth, rats, and fleas
cancer, above all diseases,
- has countless seconday causes
- primary – replacement of respiration of oxygen in normal body tissue by fermentation of glucose with conversion from obligate aerobic to anaerobic, as in bacterial cells
The cornerstone to understanding cancer is in study of the energetics of life
This thinking came out of decades of work in the Dahlem Institute Kaiser Wilhelm pre WWII and Max Planck Institute after WWII, supported by the Rockefeller Foundation.
- The oxygen- and hydrogen-transferring enzymes were discovered and isolated.
- The methods were elegant for that time, using a manometer that improved on the method used by Haldane, that did not allow the leakage of O2 or CO2.
- The interest was initiated by the increased growth of Sea Urchin eggs after fertization, which turned out to be not comparable to the rapid growth of cancer cells.
- Warburg used both normal and cancer tissue and measured the utilization of O2. He found
- that the normal tissue did not accumulate lactic acid.
- Cancer tissue generated lactic acid
- the rate of O2 consumption the same as normal tissue, but
- the rate of lactate formation far exceeded any tissue, except the retina.
- This was a discovery studied by “Pasteur” 60 years earlier (facultative aerobes), which he called the Pasteur effect.
- Hematopoietic cells of bone marrow develop aerobic glycolysis when exposed to a low oxygen condition.
He then followed on an observation by Otto Meyerhoff (Embden-Myerhoff cycle) that in muscle
- the consumption of one molecule of oxygen generates two molecules of lactate, but in aerobic glycolysis, the relationship disappears.
- He expressed the effectiveness of respiration by the ‘Meyerhoff quotient’.
- He found that cancer cells didn’t have a quotient of ‘2’
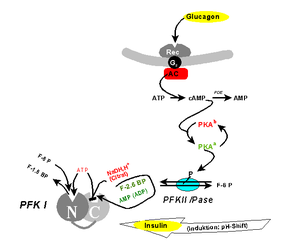
The role of the allosteric enzyme phosphofructokinase (PFK) not then known, would tie together the glycolytic and gluconeogenic pathways.
He used a heavy metal ion chelator ethylcarbylamine to
- sever the link and turn on aerobic glycolysis.
The explanation for this was provided years later by the work fleshed out by Lynen, Bucher, Lowry, Racker, and Sols.
- The rate-limiting enzyme, PFK is regulated by the concentrations of ATP, ADP, and inorganic phosphate. The ethylcarbylamide was an ‘uncoupler’ of oxidative phosphorylation.
Warburg understood that when normal cells switched to aerobic glycolysis
- it is a re-orientation of normal cell expression.
- this provides the basis for the inference that neoplastic cells become more like each other than their cell of origin.
- embryonic cells can be transformed into cancer cells under hypoxic conditions
- re-exposure to higher oxygen did not cause reversion back to normal cells.
Warburg publically expressed the rejected view in 1954 (at age 83) that restriction of chemical wastes, food additives, and air pollution would substantially reduce cancer rates.
His emphasis on the impairment of respiration was inadequate.
- the prevailing view today is loss of controlled growth of normal cells in cancer cells.
Otto Warburg: Cell Physiologist, Biochemist, and Eccentric. Hans Krebs, in collaboration with Roswitha Schmid. Clarendon Press, Oxford. 1991.ISBN 0-19-858171-8.
The Human Genome Project
The Human Genome Project, driven by Francis Collins at NIH, and by Craig Venter at the Institute for Genome Research (TIGR) had parallel projects to map the human chromosome, completed in 2003. It originally aimed to map the nucleotides contained in a human haploid reference genome (more than three billion). TIGR was the first complete genomic sequencing of a free living organism, Haemophilus influenzae, in 1995. This used a shotgun sequencing technique pioneered earlier, but which had never been used for a whole bacterium.
Venter broke away from the HGP and started Celera in 1998 because of resistance to the shotgun sequency method, and his team completed the genome sequence in three years – seven years’ less time than the HGP timetable (using the gene of Dr. Venter). TIGR eventually sequenced and analyzed more than 50 microbial genomes. Its bioinformatics group developed
- pioneering software algorithms that were used to analyze these genomes,
- including the automatic gene finder GLIMMER and
- the sequence alignment program MUMmer.
In 2002, Venter created and personally funded the J. Craig Venter Institute (JCVI) Joint Technology Center (JTC), which specialized in high throughput sequencing. The JTC, in the top ranks of scientific institutions worldwide, sequenced nearly 100 million base pairs of DNA per day for its affiliated institutions (JCVI) .
He received his his Ph.D. degree in physiology and pharmacology from the University of California, San Diego in 1975 under biochemist Nathan O. Kaplan. A full professor at the State University of New York at Buffalo, he joined the National Institutes of Health in 1984. There he learned of a technique for rapidly identifying all of the mRNAs present in a cell and began to use it to identify human brain genes. The short cDNA sequence fragments discovered by this method are called expressed sequence tags (ESTs), a name coined by Anthony Kerlavage at TIGR.
Venter believed that shotgun sequencing was the fastest and most effective way to get useful human genome data. There was a belief that shotgun sequencing was less accurate than the clone-by-clone method chosen by the HGP, but the technique became widely accepted by the scientific community and is still the de facto standard used today.
References
Shreeve, James (2004). The Genome War: How Craig Venter Tried to Capture the Code of Life and Save the World. Knopf. ISBN 0375406298.
Sulston, John (2002). The Common Thread: A Story of Science, Politics, Ethics and the Human Genome. Joseph Henry Press. ISBN 0309084091.
“The Human Genome Project Race”. Center for Biomolecular Science & Engineering, UC Santa Cruz. Retrieved 20 March 2012.
Venter, J. Craig (2007). A Life Decoded: My Genome: My Life. Viking Adult. ISBN 0670063584.
Use of a Fluorophor Probe
An article has been discussed by Dr. Tilda Barilya on use of a sensitive fluorescent probe in the near IR spectrum at > 700 nm to identify malignant ovarian cells in-vivo in abdominal exploration by tagging an overexpressed FR-α (folate-FITA)
The author makes the point that:
- In ovarian cancer, the FR-α appears to constitute a good target because it is overexpressed in 90–95% of malignant tumors, especially serous carcinomas.
- Targeting ligand, folate, is attractive as it is nontoxic, inexpensive and relatively easily conjugated to a fluorescent dye to create a tumor-specific fluorescent contrast agent.
- The report is identified as “ the first in-human proof-of-principle of the use of intraoperative tumor-specific fluorescence imaging in staging and debulking surgery for ovarian cancer using the systemically administered targeted fluorescent agent folate-FITC.”
While this does invoke possibilities for prognosis, the decision to perform the surgery, whether laparoscopic or open, is late in the discovery process. However, it does suggest the possibility that the discovery and the treatment might be combined if the biomarker itself had the fluorescence to identify the overexpression, but it also is combined with a tag to block the overexpession. This hypothetical possibility is now expressed below.
http://pharmaceuticalintelligence.com/2013/01/19/ovarian-cancer-and-fluorescence-guided-surgery-a-report/
Gene Editing
Dr. Aviva Lev-Ari reports that a new technique developed at MIT Broad Institute and the Rockefeller University can edit DNA in precise locations taken from Science News titled “Editing Genome With High Precision: New Method to Insert Multiple Genes in Specific Locations, Delete Defective Genes”.
Using this system, scientists can alter
- several genome sites simultaneously and
- can achieve much greater control over where new genes are inserted
According to Feng Zhang, this is an improvement beyond splicing the gene in specific locations and insertion of complexes difficult to assemble known as transcription activator-like effector nucleases (TALENs).
- The researchers create DNA-editing complexes
- using naturally occurring bacterial protein-RNA systems
- that recognize and snip viral DNA, including
- a nuclease called Cas9 bound to short RNA sequences.
- they target specific locations in the genome, and
- when they encounter a match, Cas9 cuts the DNA.
This approach can be used either to
- disrupt the function of a gene or
- to replace it with a new one.
- To replace the gene, a DNA template for the new gene has to be copied into the genome after the DNA is cut. The method is also very precise —
- if there is a single base-pair difference between the RNA targeting sequence and the genome sequence, Cas9 is not activated.
In its first iteration, it appears comparable in efficiency to what zinc finger nucleases and TALENs have to offer.
The research team has deposited the necessary genetic components with a nonprofit called Addgene, and they have also created a website with tips and tools for using this new technique.
The above story is reprinted from materials provided by Massachusetts Institute of Technology. The original article was written by Anne Trafton.
Le Cong, F. Ann Ran, David Cox, Shuailiang Lin, Robert Barretto, Naomi Habib, Patrick D. Hsu, Xuebing Wu, Wenyan Jiang, Luciano Marraffini, and Feng Zhang. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science, 3 January 2013 DOI: 10.1126/science.1231143. http://Science.com. Editing genome with high precision: New method to insert multiple genes in specific locations, delete defective genes. ScienceDaily. Retrieved January 20, 2013, from http://www.sciencedaily.com /releases/2013/01/130103143205.htm?goback=%2Egde_4346921_member_205356312.
Dr. Lev-Ari also reports on a study of early detection of breast cancer in “Mechanism involved in Breast Cancer Cell Growth: Function in Early Detection & Treatment“, by Dr. Rotem Karni and PhD student Vered Ben Hur at the Institute for Medical Research Israel-Canada of the Hebrew University. http://pharmaceuticalintelligence.com/2013/01/17/mechanism-involved-in-breast-cancer-cell-growth-function-in-early-detection-treatment/
These researchers have discovered a new mechanism by which breast cancer cells switch on their aggressive cancerous behavior. The discovery provides a valuable marker for the early diagnosis and follow-up treatment of malignant growths.
The method they use is
- RNA splicing and insertion.
- The information needed for the production of a mature protein is encoded in segments called exons .
- In the splicing process, the non-coding segments of the RNA (introns) are spliced from the pre-mRNA and
- the exons are joined together.
Alternative splicing is when a specific ”scene” (or exon) is either inserted or deleted from the movie (mRNA), thus changing its meaning.
- Over 90 percent of the genes in our genome undergo alternative splicing of one or more of their exons, and
- the resulting changes in the proteins encoded by these different mRNAs are required for normal function.
- the normal process of alternative splicing is altered in cancer, and
- ”bad” protein forms are generated that aid cancer cell proliferation and survival.
The researchers reported in online Cell Reports that breast cancer cells
- change the alternative splicing of an important enzyme, called S6K1, which is
- a protein involved in the transmission of information into the cell.
- when this happens, breast cancer cells start to produce shorter versions of this enzyme and
- these shorter versions transmit signals ordering the cells to grow, proliferate, survive and invade other tissues (otherwise proliferation is suppressed)
The application to biotherapeutics would be to ”reverse” the alternative splicing of S6K1 in cancer cells back to the normal situation as a novel anti-cancer therapy.
Additional Developments:
A*STAR Scientists Pinpoint Genetic Changes that Spell Cancer: Fruit flies light the way for scientists to uncover genetic changes.
With a new approach, researchers may rapidly distinguish the range of
- genetic changes that are causally linked to cancer (i.e. “driver” mutations)
- versus those with limited impact on cancer progression.
This study published in the prestigious journal Genes & Development could pave the way to design more targeted treatment against different cancer types, based on the specific cancer-linked mutations present in the patient, an advance in the development of personalized medicine.
Signaling pathways involved in tumour formation are conserved from fruit flies to humans. In fact, about 75 percent of known human disease genes have a recognizable match in the genome of fruit flies.
Leveraging on their genetic similarities, Dr Hector Herranz, a post-doctorate from the Dr. Stephen Cohen’s team developed an innovative strategy to genetically screen the whole fly genome for “cooperating” cancer genes.
- These genes appear to have little or no impact on cancer.
- However, they cooperate with other cancer genes, so that
- the combination causes aggressive cancer, which
- neither would cause alone.
In this study, the team was specifically looking for genes that
- could cooperate with EGFR “driver” mutation,
- a genetic change commonly associated with breast and lung cancers in humans.
- SOCS5 (reported in this paper) is one of the several new “cooperating” cancer genes to be identified.
Already, there are indications that levels of SOCS5 expression are
- reduced in breast cancer, and
- patients with low levels of SOCS5 have poor prognosis.”
The IMCB team is preparing to explore the use of SOCS5 as a biomarker in diagnosis for cancer.
http://genes&development.com
Probing What Fuels Cancer
‘Altered cellular metabolism is a hallmark of cancer,’ says Dr Patrick Pollard, in the Nuffield Department of Clinical Medicine at Oxford. Most cancer cells get the energy they need predominantly through a high rate of glycolysis, allowing cancer cells deal with the low oxygen levels that tend to be present in a tumour.
But whether dysfunctional metabolism causes cancer, as Warburg believed, or is something that happens afterwards is a different question. In the meantime, gene studies rapidly progressed and indicated that genetic changes occur in cancer.
DNA mutations spring up all the time in the body’s cells, but
- most are quickly repaired.
- Alternatively the cell might shut down or be killed off (apoptosis) before any damage is caused. However, the repair machinery is not perfect.
- If changes occur that bypass parts of the repair machinery or sabotage it,
- the cell can escape the body’s normal controls on growth and
- DNA changes can begin to accumulate as the cell becomes cancerous.
Patrick believes certain changes in cells can’t always be accounted for by ‘genetics.’
He is now collaborating with Professor Tomoyoshi Soga’s large lab at Keio University in Japan, which has been at the forefront of developing the technology for metabolomics research over the past couple of decades.
The Japanese lab’s ability to
- screen samples for thousands of compounds and metabolites at once, and
- the access to tumour material and cell and animal models of disease
- enables them to probe the metabolic changes that occur in cancer.
There is reason to believe that
- dysfunctional cell metabolism is important in cancer.
- genes with metabolic functions are associated with some cancers
- changes in the function of a metabolic enzyme have been implicated in the development of gliomas.
These results have led to the idea that some metabolic compounds, or metabolites, when they accumulate in cells, can cause changes to metabolic processes and set cells off on a path towards cancer.
Patrick Pollard and colleagues have now published a perspective article in the journal Frontiers in Molecular and Cellular Oncology that proposes fumarate as such an ‘oncometabolite’. Fumarate is a standard compound involved in cellular metabolism.
The researchers summarize evidence that shows how
- accumulation of fumarate when an enzyme goes wrong affects various biological pathways in the cell.
- It shifts the balance of metabolic processes and disrupts the cell in ways that could favour development of cancer.
Patrick and colleagues write in their latest article that the shift in focus of cancer research to include cancer cell metabolism ‘has highlighted how woefully ignorant we are about the complexities and interrelationships of cellular metabolic pathways’.
http://FrontiersMolecularCellularOncology.com
Using genome-wide Chromatin Interaction Analysis with Paired-End-Tag sequencing (ChIA-PET),
mapped long-range chromatin interactions associated with RNA polymerase II in human cells
uncovered widespread promoter-centered intragenic, extragenic, and intergenic interactions.
- These interactions further aggregated into higher-order clusters
- proximal and distal genes were engaged through promoter-promoter interactions.
- most genes with promoter-promoter interactions were active and transcribed cooperatively
- some interacting promoters could influence each other implying combinatorial complexity of transcriptional controls.
Comparative analyses of different cell lines showed that
- cell-specific chromatin interactions could provide structural frameworks for cell-specific transcription,
- and suggested significant enrichment of enhancer-promoter interactions for cell-specific functions.
- genetically-identified disease-associated noncoding elements were spatially engaged with corresponding genes through long-range interactions.
Overall, our study provides insights into transcription regulation by
- three-dimensional chromatin interactions for both housekeeping and
- cell-specific genes in human cells.
New Nucleoporin: Regulator of Transcriptional Repression and Beyond.
(NJ Sarma and K Willis) Nucleus 2012; 3(6): 1–8; http://Nucleus.com © 2012 Landes Bioscience
Transcriptional regulation is a complex process that requires the integrated action of many multi-protein complexes.
The way in which a living cell coordinates the action of these complexes in time and space is still poorly understood.
- nuclear pores, well known for their role in 3′ processing and export of transcripts, also participate in the control of transcriptional initiation.
- nuclear pores interface with the well-described machinery that regulates initiation.
This work led to the discovery that
- specific nucleoporins are required for binding of the repressor protein Mig1 to its site in target promoters.
- Nuclear pores are involved in repressing, as well as activating, transcription.
Here we discuss in detail the main models explaining our result and consider what each implies about the roles that nuclear pores play in the regulation of gene expression.
Prediction of Breast Cancer Metastasis by Gene Expression Profiles: A Comparison of Metagenes and Single Genes.
(M Burton, M Thomassen, Q Tan, and TA Kruse.) Cancer Informatics 2012:11 193–217 doi: 10.4137/CIN.S10375
The popularity of a large number of microarray applications has in cancer research led to the development of predictive or prognostic gene expression profiles. However, the diversity of microarray platforms has made the full validation of such profiles and their related gene lists across studies difficult and, at the level of classification accuracies, rarely validated in multiple independent datasets. Frequently, while the individual genes between such lists may not match, genes with same function are included across such gene lists. Development of such lists does not take into account the fact that
- genes can be grouped together as metagenes (MGs) based on common characteristics such as pathways, regulation, or genomic location.
In this study we compared the performance of either metagene- or single gene-based feature sets and classifiers using random forest and two support vector machines for classifier building. The performance
- within the same dataset,
- feature set validation performance, and
- validation performance of entire classifiers in strictly independent datasets
were assessed by
- 10 times repeated 10-fold cross validation,
- leave-one-out cross validation, and
- one-fold validation, respectively.
To test the significance of the performance difference between MG- and SG-features/classifiers, we used a repeated down-sampled binomial test approach.
MG- and SG-feature sets are transferable and perform well for training and testing prediction of metastasis outcome
- in strictly independent data sets, both
- between different and
- within similar microarray platforms, while
- classifiers had a poorer performance when validated in strictly independent datasets.
The study showed that MG- and SG-feature sets perform equally well in classifying independent data. Furthermore, SG-classifiers significantly outperformed MG-classifier
- when validation is conducted between datasets using similar platforms, while
- no significant performance difference was found when validation was performed between different platforms.
Prediction of metastasis outcome in lymph node–negative patients by MG- and SG-classifiers showed that SG-classifiers performed significantly better than MG-classifiers when validated in independent data based on the same microarray platform as used for developing the classifier. However, the MG- and SG-classifiers had similar performance when conducting classifier validation in independent data based on a different microarray platform. The latter was also true when only validating sets of MG- and SG-features in independent datasets, both between and within similar and different platforms.
Identification and Insilico Analysis of Retinoblastoma Serum microRNA Profile and Gene Targets Towards Prediction of Novel Serum Biomarkers.
M Beta, A Venkatesan, M Vasudevan, U Vetrivel, et al. Identification and Insilico Analysis of Retinoblastoma Serum microRNA Profile and Gene Targets Towards Prediction of Novel Serum Biomarkers.
http://Bioinformatics and Biology Insights 2013:7 21–34. doi: 10.4137/BBI.S10501
This study was undertaken
- to identify the differentially expressed miRNAs in the serum of children with RB in comparison with the normal age matched serum,
- to analyze its concurrence with the existing RB tumor miRNA profile,
- to identify its novel gene targets specific to RB, and
- to study the expression of a few of the identified oncogenic miRNAs in the advanced stage primary RB patient’s serum sample.
MiRNA profiling performed on 14 pooled serum from children with advanced RB and 14 normal age matched serum samples
- 21 miRNAs found to be upregulated (fold change > 2.0, P < 0.05) and
- 24 downregulated (fold change > 2.0, P < 0.05).
Intersection of 59 significantly deregulated miRNAs identified from RB tumor profiles with that of miRNAs detected in serum profile revealed that
- 33 miRNAs had followed a similar deregulation pattern in RB serum.
Later we validated a few of the miRNAs (miRNA 17-92) identified by microarray in the RB patient serum samples (n = 20) by using qRT-PCR.
Expression of the oncogenic miRNAs, miR-17, miR-18a, and miR-20a by qRT-PCR was significant in the serum samples
- exploring the potential of serum miRNAs identification as noninvasive diagnosis.
Moreover, from miRNA gene target prediction, key regulatory genes of
- cell proliferation,
- apoptosis, and
- positive and negative regulatory networks
involved in RB progression were identified in the gene expression profile of RB tumors.
Therefore, these identified miRNAs and their corresponding target genes could give insights on
- potential biomarkers and key events involved in the RB pathway.
Computational Design of Targeted Inhibitors of Polo-Like Kinase 1 ( lk1).
(KS Jani and DS Dalafave) Bioinformatics and Biology Insights 2012:6 23–31.doi: 10.4137/BBI.S8971.
Computational design of small molecule putative inhibitors of Polo-like kinase 1 (Plk1) is presented. Plk1, which regulates the cell cycle, is often over expressed in cancers.
- Down regulation of Plk1 has been shown to inhibit tumor progression.
- Most kinase inhibitors interact with the ATP binding site on Plk1, which is highly conserved.
- This makes the development of Plk1-specific inhibitors challenging, since different kinases have similar ATP sites.
However, Plk1 also contains a unique region called the polo-box domain (PBD), which is absent from other kinases.
- the PBD site was used as a target for designed Plk1 putative inhibitors.
- Common structural features of several experimentally known Plk1 ligands were first identified.
- The findings were used to design small molecules that specifically bonded Plk1.
- Drug likeness and possible toxicities of the molecules were investigated.
- Molecules with no implied toxicities and optimal drug likeness values were used for docking studies.
- Several molecules were identified that made stable complexes only with Plk1 and LYN kinases, but not with other kinases.
- One molecule was found to bind exclusively the PBD site of Plk1.
Possible utilization of the designed molecules in drugs against cancers with over expressed Plk1 is discussed.
Conclusions
The previous discussions reviewed the status of an evolving personalized medicine multicentered and worldwide enterprise. It is also clear from these reports that the search for targeted drugs matched to a cancer profile or signature has identified several approaches that show great promise.
- We know considerably more about metabolic pathways and linked changes in transcription that occur in neoplastic development.
- There are several methods used to do highly accurate insertions in gene sequences that are linked to specific metabolic changes, and
- some may have significant implications for therapeutics, if
- the link is a change that is associated with a driver mutation
- the link can be identified by a fluorescent or other probe
- the link is tied to a mRNA or peptide product that is a biomarker measured in the circulation
- We have probes to genetic links to the control of many and interacting signaling pathways.
- We know more about transcription through mRNA.
- We are closer to the possibility that metabolic substrates, like ‘fumarate’ (a key intermediate in the TCA cycle), may provide a means to reverse regulate the neoplastic cells.
- We may also find metabolic channels that drive the cells from proliferation to apoptosis or normal activity.
Summary
This discussion identified the huge expansion of genomic technology in the investigation of biopharmacotherapeutic targets that have been identified involving different levels and interacting signaling pathways. There are several methods of analyzing gene expression, and a primary emphasis is given to combinations of mutations expressed in different cancer types. There is a major hypothesis that expresses the need to focus on “central” “driver mutations” that correspond with the regulation of gene expression, cell proliferation, and cell metabolism. What hasn’t been know is why drug resistance develops and whether the cellular migration and aerobic glycolysis can be redirected after cell metastasis occurs.
.

A slight mutation in the matched nucleotides can lead to chromosomal aberrations and unintentional genetic rearrangement. (Photo credit: Wikipedia)
Additional Related articles
- Unraveling the Human Genome: 6 Molecular Milestones (livescience.com)
- The Role Of Microorganisms In Cancer Is Being Ignored By The Current Sequencing Strategies (3quarksdaily.com)
- Biological Functions of Noncoding DNA (tginnovations.wordpress.com)
- Retrovirus in the human genome is active in pluripotent stem cells (sciencedaily.com)
- Consistent Personal Epigenetic ‘Signatures’ Discovered In Prostate Cancer Patients’ Metastases (medicalnewstoday.com)
- Melanomas Often Mutate in Two Specific Ways, Shows Study (webpronews.com)
Other posts related to this discussion were published on this Open Source Online Scientific Journal from Leaders in Pharmaceutical Business Intelligence:
Big Data in Genomic Medicine, LHB
http://pharmaceuticalintelligence.com/2012/12/17/big-data-in-genomic-medicine/
A New Therapy for Melanoma, LHB
http://pharmaceuticalintelligence.com/2012/09/15/a-new-therapy-for-melanoma/
BRCA1 a tumour suppressor in breast and ovarian cancer – functions in transcription, ubiquitination and DNA repair, S Saha
http://pharmaceuticalintelligence.com/2012/12/04/brca1-a-tumour-suppressor-in-breast-and-ovarian-cancer-functions-in-transcription-ubiquitination-and-dna-repair/
Judging ‘Tumor response’-there is more food for thought, R Saxena
http://pharmaceuticalintelligence.com/2012/12/04/judging-the-tumor-response-there-is-more-food-for-thought/
Computational Genomics Center: New Unification of Computational Technologies at Stanford, A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/12/03/computational-genomics-center-new-unification-of-computational-technologies-at-stanford/
Ovarian Cancer and fluorescence-guided surgery: A report, T. Barliya
http://pharmaceuticalintelligence.com/2013/01/19/ovarian-cancer-and-fluorescence-guided-surgery-a-report/
Personalized medicine gearing up to tackle cancer , R. Saxena
http://pharmaceuticalintelligence.com/2013/01/07/personalized-medicine-gearing-up-to-tackle-cancer/
Exploring the role of vitamin C in Cancer therapy, R. Saxena
http://pharmaceuticalintelligence.com/2013/01/15/exploring-the-role-of-vitamin-c-in-cancer-therapy/
Differentiation Therapy – Epigenetics Tackles Solid Tumors, SJ Williams
http://pharmaceuticalintelligence.com/2013/01/03/differentiation-therapy-epigenetics-tackles-solid-tumors/
Mechanism involved in Breast Cancer Cell Growth: Function in Early Detection & Treatment, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/17/mechanism-involved-in-breast-cancer-cell-growth-function-in-early-detection-treatment/
Personalized Medicine: Cancer Cell Biology and Minimally Invasive Surgery (MIS), A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/12/01/personalized-medicine-cancer-cell-biology-and-minimally-invasive-surgery-mis/
Role of Primary Cilia in Ovarian Cancer, A. Awan
http://pharmaceuticalintelligence.com/2013/01/15/role-of-primary-cilia-in-ovarian-cancer-2/
The Molecular Pathology of Breast Cancer Progression, T. Bailiya`
http://pharmaceuticalintelligence.com/2013/01/10/the-molecular-pathology-of-breast-cancer-progression/
Stanniocalcin: A Cancer Biomarker, A. Awan
http://pharmaceuticalintelligence.com/2012/12/25/stanniocalcin-a-cancer-biomarker/
Nanotechnology, personalized medicine and DNA sequencing, T. Barliya
http://pharmaceuticalintelligence.com/2013/01/09/nanotechnology-personalized-medicine-and-dna-sequencing/
Gastric Cancer: Whole-genome reconstruction and mutational signatures, A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/12/24/gastric-cancer-whole-genome-reconstruction-and-mutational-signatures-2/
Paradigm Shift in Human Genomics – Predictive Biomarkers and Personalized Medicine – Part 1, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/paradigm-shift-in-human-genomics-predictive-biomarkers-and-personalized-medicine-part-1/
LEADERS in Genome Sequencing of Genetic Mutations for Therapeutic Drug Selection in Cancer Personalized Treatment: Part 2, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/leaders-in-genome-sequencing-of-genetic-mutations-for-therapeutic-drug-selection-in-cancer-personalized-treatment-part-2/
Personalized Medicine: An Institute Profile – Coriell Institute for Medical Research: Part 3, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/personalized-medicine-an-institute-profile-coriell-institute-for-medical-research-part-3/
The Consumer Market for Personal DNA Sequencing: Part 4, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/consumer-market-for-personal-dna-sequencing-part-4/
Harnessing Personalized Medicine for Cancer Management, Prospects of Prevention and Cure: Opinions of Cancer Scientific Leaders @ http://pharmaceuticalintelligence.com A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/13/7000/
GSK for Personalized Medicine using Cancer Drugs needs Alacris systems biology model to determine the in silico effect of the inhibitor in its “virtual clinical trial” A Lev-Ari
http://pharmaceuticalintelligence.com/2012/11/14/gsk-for-personalized-medicine-using-cancer-drugs-needs-alacris-systems-biology-model-to-determine-the-in-silico-effect-of-the-inhibitor-in-its-virtual-clinical-trial/
Recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes in serous endometrial tumors, S. Saha
http://pharmaceuticalintelligence.com/2012/11/19/recurrent-somatic-mutations-in-chromatin-remodeling-and-ubiquitin-ligase-complex-genes-in-serous-endometrial-tumors/
Metabolic drivers in aggressive brain tumors, pkandala
http://pharmaceuticalintelligence.com/2012/11/11/metabolic-drivers-in-aggressive-brain-tumors/
Personalized medicine-based cure for cancer might not be far away, R. Saxena
http://pharmaceuticalintelligence.com/2012/11/20/personalized-medicine-based-cure-for-cancer-might-not-be-far-away/
Response to Multiple Cancer Drugs through Regulation of TGF-β Receptor Signaling: a MED12 Control, A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/11/21/response-to-multiple-cancer-drugs-through-regulation-of-tgf-%CE%B2-receptor-signaling-a-med12-control/
Human Variome Project: encyclopedic catalog of sequence variants indexed to the human genome sequence, A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/11/24/human-variome-project-encyclopedic-catalog-of-sequence-variants-indexed-to-the-human-genome-sequence/
Prostate Cancer Cells: Histone Deacetylase Inhibitors Induce Epithelial-to-Mesenchymal Transition, SJ Williams
http://pharmaceuticalintelligence.com/2012/11/30/histone-deacetylase-inhibitors-induce-epithelial-to-mesenchymal-transition-in-prostate-cancer-cells/
Tumor Imaging and Targeting: Predicting Tumor Response to Treatment: Where we stand?, R. Saxena
http://pharmaceuticalintelligence.com/2012/12/13/imaging-and-targeting-the-tumor-predicting-tumor-response-where-we-stand/
Nanotechnology: Detecting and Treating metastatic cancer in the lymph node, T. Barliya
http://pharmaceuticalintelligence.com/2012/12/19/nanotechnology-detecting-and-treating-metastatic-cancer-in-the-lymph-node/
Heroes in Medical Research: Barnett Rosenberg and the Discovery of Cisplatin, SJ Williams
http://pharmaceuticalintelligence.com/2013/01/12/heroes-in-medical-research-barnett-rosenberg-and-the-discovery-of-cisplatin/
Inspiration From Dr. Maureen Cronin’s Achievements in Applying Genomic Sequencing to Cancer Diagnostics, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/10/inspiration-from-dr-maureen-cronins-achievements-in-applying-genomic-sequencing-to-cancer-diagnostics/
The “Cancer establishments” examined by James Watson, co-discoverer of DNA w/Crick, 4/1953, A. Lev-Ari
http://pharmaceuticalintelligence.com/2013/01/09/the-cancer-establishments-examined-by-james-watson-co-discover-of-dna-wcrick-41953/
Nanotech Therapy for Breast Cancer. T. Barlyia
http://pharmaceuticalintelligence.com/2012/12/09/naotech-therapy-for-breast-cancer/
Dasatinib in Combination With Other Drugs for Advanced, Recurrent Ovarian Cancer, pkandala
http://pharmaceuticalintelligence.com/2012/12/08/dasatinib-in-combination-with-other-drugs-for-advanced-recurrent-ovarian-cancer/
Squeezing Ovarian Cancer Cells to Predict Metastatic Potential: Cell Stiffness as Possible Biomarker, pkandala
http://pharmaceuticalintelligence.com/2012/12/08/squeezing-ovarian-cancer-cells-to-predict-metastatic-potential-cell-stiffness-as-possible-biomarker/
Hypothesis – following on James Watson, LHB
http://pharmaceuticalintelligence.com/2013/01/27/novel-cancer-h…ts-are-harmful/
Otto Warburg, A Giant of Modern Cellular Biology, LHB
http://pharmaceuticalintelligence.com/2012/11/02/otto-warburg-a-giant-of-modern-cellular-biology/
Is the Warburg Effect the cause or the effect of cancer: A 21st Century View? LHB
http://pharmaceuticalintelligence.com/2012/10/17/is-the-warburg-effect-the-cause-or-the-effect-of-cancer-a-21st-century-view/
Remembering a Great Scientist among Mentors, LHB
http://pharmaceuticalintelligence.com/2013/01/26/remembering-a-great-scientist-among-mentors/
Portrait of a great scientist and mentor: Nathan Oram Kaplan, LHB
http://pharmaceuticalintelligence.com/2013/01/26/portrait-of-a-great-scientist-and-mentor-nathan-oram-kaplan/
Predicting Tumor Response, Progression, and Time to Recurrence, LHB
http://pharmaceuticalintelligence.com/2012/12/20/predicting-tumor-response-progression-and-time-to-recurrence/
Directions for genomics in personalized medicine, LHB
http://pharmaceuticalintelligence.com/2013/01/27/directions-for-genomics-in-personalized-medicine/
How mobile elements in “Junk” DNA promote cancer. Part 1: Transposon-mediated tumorigenesis, Sjwilliams
http://pharmaceuticalintelligence.com/2012/10/31/how-mobile-elements-in-junk-dna-prote-cacner-part1-transposon-mediated-tumorigenesis/
Novel Cancer Hypothesis Suggests Antioxidants Are Harmful, LHB
http://pharmaceuticalintelligence.com/2013/01/27/novel-cancer-hypothesis-suggests-antioxidants-are-harmful/
Mitochondria: Origin from oxygen free environment, role in aerobic glycolysis, metabolic adaptation, LHB
http://pharmaceuticalintelligence.com/2012/09/26/mitochondria-origin-from-oxygen-free-environment-role-in-aerobic-glycolysis-metabolic-adaptation/
Advances in Separations Technology for the “OMICs” and Clarification of Therapeutic Targets, LHB
http://pharmaceuticalintelligence.com/2012/10/22/advances-in-separations-technology-for-the-omics-and-clarification-of-therapeutic-targets/
Cancer Innovations from across the Web, LHB
http://pharmaceuticalintelligence.com/2012/11/02/cancer-innovations-from-across-the-web/
Mitochondrial Damage and Repair under Oxidative Stress, LHB
http://pharmaceuticalintelligence.com/2012/10/28/mitochondrial-damage-and-repair-under-oxidative-stress/
Mitochondria: More than just the “powerhouse of the cell” R. Saxena
http://pharmaceuticalintelligence.com/2012/07/09/mitochondria-more-than-just-the-powerhouse-of-the-cell/
Mitochondria and Cancer: An overview of mechanisms, R. Saxena
http://pharmaceuticalintelligence.com/2012/09/01/mitochondria-and-cancer-an-overview/
Mitochondrial fission and fusion: potential therapeutic targets? R. Saxena
http://pharmaceuticalintelligence.com/2012/10/31/mitochondrial-fission-and-fusion-potential-therapeutic-target/
Mitochondrial mutation analysis might be “1-step” away, R. Saxena
http://pharmaceuticalintelligence.com/2012/08/14/mitochondrial-mutation-analysis-might-be-1-step-away/
β Integrin emerges as an important player in mitochondrial dysfunction associated Gastric Cancer, R. Saxena
http://pharmaceuticalintelligence.com/2012/09/10/%CE%B2-integrin-emerges-as-an-important-player-in-mitochondrial-dysfunction-associated-gastric-cancer/
mRNA interference with cancer expression, LHB
http://pharmaceuticalintelligence.com/2012/10/26/mrna-interference-with-cancer-expression/
What can we expect of tumor therapeutic response? LHB
http://pharmaceuticalintelligence.com/2012/12/05/what-can-we-expect-of-tumor-therapeutic-response/
Expanding the Genetic Alphabet and linking the genome to the metabolome, LHB
http://pharmaceuticalintelligence.com/2012/09/24/expanding-the-genetic-alphabet-and-linking-the-genome-to-the-metabolome/
Breast Cancer, drug resistance, and biopharmaceutical targets, LHB
http://pharmaceuticalintelligence.com/2012/09/18/breast-cancer-drug-resistance-and-biopharmaceutical-targets/
Breast Cancer: Genomic Profiling to Predict Survival: Combination of Histopathology and Gene Expression Analysis, A. Lev-Ari
http://pharmaceuticalintelligence.com/2012/12/24/breast-cancer-genomic-profiling-to-predict-survival-combination-of-histopathology-and-gene-expression-analysis/
Ubiquinin-Proteosome pathway, autophagy, the mitochondrion, proteolysis and cell apoptosis, LHB
http://pharmaceuticalintelligence.com/2012/10/30/ubiquinin-proteosome-pathway-autophagy-the-mitochondrion-proteolysis-and-cell-apoptosis/
Identification of Biomarkers that are Related to the Actin Cytoskeleton, LHB
http://pharmaceuticalintelligence.com/2012/12/10/identification-of-biomarkers-that-are-related-to-the-actin-cytoskeleton/
Nitric Oxide has a ubiquitous role in the regulation of glycolysis -with a concomitant influence on mitochondrial function, LHB
http://pharmaceuticalintelligence.com/2012/09/16/nitric-oxide-has-a-ubiquitous-role-in-the-regulation-of-glycolysis-with-a-concomitant-influence-on-mitochondrial-function/
Genomic Analysis: FLUIDIGM Technology in the Life Science and Agricultural Biotechnology, A. Lev-Ari http://pharmaceuticalintelligence.com/2012/08/22/genomic-analysis-fluidigm-technology-in-the-life-science-and-agricultural-biotechnology/
Nanotechnology: Detecting and Treating metastatic cancer in the lymph node, T. Barliya
http://pharmaceuticalintelligence.com/2012/12/19/nanotechnology-detecting-and-treating-metastatic-cancer-in-the-lymph-node/