
Cancer detection and therapeutics
Curator: Larry H. Bernstein, MD, FCAP
Kurzweill Reports
http://kurzweilai.us1.list-manage.com/track/click
Machine learning rivals human skills in cancer detection
Two announcements yesterday (April 21) suggest that deep learning algorithms rival human skills in detecting cancer from ultrasound images and in identifying cancer in pathology reports.

Samsung Medison RS80A ultrasound imaging system (credit: Samsung)
Samsung Medison, a global medical equipment company and an affiliate of Samsung Electronics, has just updated its RS80A ultrasound imaging system with a deep learning algorithm for breast-lesion analysis.
The “S-Detect for Breast” feature uses big data collected from breast-exam cases and recommends whether the selected lesion is benign or malignant. It’s used in in lesion segmentation, characteristic analysis, and assessment processes, providing “more accurate results.”
“We saw a high level of conformity from analyzing and detecting lesion in various cases by using the S-Detect,” said professor Han Boo Kyung, a radiologist at Samsung Medical Center.
“Users can reduce taking unnecessary biopsies and doctors-in-training will likely have more reliable support in accurately detecting malignant and suspicious lesions.”
Deep learning is better than humans in extracting meaning from cancer pathology reports
Meanwhile, researchers from the Regenstrief Institute and Indiana University School of Informatics and Computing at Indiana University-Purdue University Indianapolis say they’ve found that open-source machine learning tools are as good as — or better than — humans in extracting crucial meaning from free-text (unstructured) pathology reports and detecting cancer cases. The computer tools are also faster and less resource-intensive.
(U.S. states require cancer cases to be reported to statewide cancer registries for disease tracking, identification of at-risk populations, and recognition of unusual trends or clusters. This free-text information can be difficult for health officials to interpret, which can further delay health department action, when action is needed.)
“We think that its no longer necessary for humans to spend time reviewing text reports to determine if cancer is present or not,” said study senior author Shaun Grannis*, M.D., M.S., interim director of the Regenstrief Center of Biomedical Informatics.
Awash in oceans of data
“We have come to the point in time that technology can handle this. A human’s time is better spent helping other humans by providing them with better clinical care. Everything — physician practices, health care systems, health information exchanges, insurers, as well as public health departments — are awash in oceans of data. How can we hope to make sense of this deluge of data? Humans can’t do it — but computers can.”
This is especially relevant for underserved nations, where a majority of clinical data is collected in the form of unstructured free text, he said.
The researchers sampled 7,000 free-text pathology reports from over 30 hospitals that participate in the Indiana Health Information Exchange and used open source tools, classification algorithms, and varying feature selection approaches to predict if a report was positive or negative for cancer. The results indicated that a fully automated review yielded results similar or better than those of trained human reviewers, saving both time and money.
Major infrastructure advance
“We found that artificial intelligence was as least as accurate as humans in identifying cancer cases from free-text clinical data. For example the computer ‘learned’ that the word ‘sheet’ or ‘sheets’ signified cancer as ‘sheet’ or ‘sheets of cells’ are used in pathology reports to indicate malignancy.
“This is not an advance in ideas; it’s a major infrastructure advance — we have the technology, we have the data, we have the software from which we saw accurate, rapid review of vast amounts of data without human oversight or supervision.”
The study was published in the April 2016 issue of the Journal of Biomedical Informatics. It was conducted with support from the Centers for Disease Control and Prevention.
Co-authors of the study include researchers at the IU Fairbanks School of Public Health, the IU School of Medicine and the School of Science at IUPUI.
* Grannis, a Regenstrief Institute investigator and an associate professor of family medicine at the IU School of Medicine, is the architect of the Regenstrief syndromic surveillance detector for communicable diseases and led the technical implementation of Indiana’s Public Health Emergency Surveillance System — one of the nation’s largest. Studies over the past decade have shown that this system detects outbreaks of communicable diseases seven to nine days earlier and finds four times as many cases as human reporting while providing more complete data.
Yann Lecun is Director of AI Research, Facebook and a noted deep-learning expert.
Toward better public health reporting using existing off the shelf approaches: A comparison of alternative cancer detection approaches using plaintext medical data and non-dictionary based feature selection
Suranga N. Kasthurirathnea, , , Brian E. Dixonb, c, Judy Gichoyad, Huiping Xuc, Yuni Xiad, Burke Mamlinb, d, Shaun J. Grannisb, d
Highlights
- • Cancer cases can be identified in unstructured clinical data to support public health reporting.
- • Such cancer detection methods do not require complex external ontologies or human intervention.
- • Such approaches can identify cases with sensitivity, specificity, PPV, accuracy, and AUC exceeding 80–90%.
- • Automated cancer detection methods perform as well as approaches that require costly clinician input.
- • These approaches may be generalized for other health analytics applications and healthcare domains.
Abstract
Objectives
Increased adoption of electronic health records has resulted in increased availability of free text clinical data for secondary use. A variety of approaches to obtain actionable information from unstructured free text data exist. These approaches are resource intensive, inherently complex and rely on structured clinical data and dictionary-based approaches. We sought to evaluate the potential to obtain actionable information from free text pathology reports using routinely available tools and approaches that do not depend on dictionary-based approaches.
Materials and methods
We obtained pathology reports from a large health information exchange and evaluated the capacity to detect cancer cases from these reports using 3 non-dictionary feature selection approaches, 4 feature subset sizes, and 5 clinical decision models: simple logistic regression, naïve bayes, k-nearest neighbor, random forest, and J48 decision tree. The performance of each decision model was evaluated using sensitivity, specificity, accuracy, positive predictive value, and area under the receiver operating characteristics (ROC) curve.
Results
Decision models parameterized using automated, informed, and manual feature selection approaches yielded similar results. Furthermore, non-dictionary classification approaches identified cancer cases present in free text reports with evaluation measures approaching and exceeding 80–90% for most metrics.
Conclusion
Our methods are feasible and practical approaches for extracting substantial information value from free text medical data, and the results suggest that these methods can perform on par, if not better, than existing dictionary-based approaches. Given that public health agencies are often under-resourced and lack the technical capacity for more complex methodologies, these results represent potentially significant value to the public health field.

Scientists shoot anticancer drugs deep into tumors
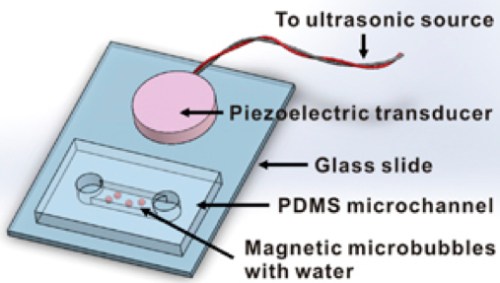
Schematic of a magnetic microbubble used in the study, containing gas core (blue) and shell of magnetic iron-oxide nanoparticles (red) that form a dense shell (center) around drug-containing nanoparticles. When stimulated by ultrasound at resonant frequencies, the microbubbles explode, releasing the nanoparticles, which can travel hundreds of micrometers into tumor tissue to deliver anticancer drugs and can also be imaged on an MRI machine. (credit: Yu Gao et al./NPG Asia Materials) http://www.kurzweilai.net/images/magnetic-microbubble.jpg
Scientists at Nanyang Technological University (NTU Singapore) have invented a new way to deliver cancer drugs deep into tumor cells.
They created micro-sized gas bubbles coated with anticancer drug particles embedded in iron oxide nanoparticles and used MRI or other magnetic sources to direct these microbubbles to gather around a specific tumor. Then they used ultrasound to vibrate the microbubbles, providing the energy to direct the drug particles into a targeted kill zone in the tumor. The magnetic nanoparticles also allow for imaging in an MRI machine.
The microbubbles were successfully tested in mice and the study has been published by the Nature Publishing Group in Asia Materials.
Overcoming limitations of chemotherapy
This innovative technique was developed by a multidisciplinary team of scientists led by Asst Prof C. J. Xu from the School of Chemical and Biomedical Engineering and Assoc. Prof Claus-Dieter Ohl from the School of Physical and Mathematical Sciences.
Xu, who is also a researcher at the NTU-Northwestern Institute for Nanomedicine, said their new method may solve some of the most pressing problems faced in chemotherapy used to treat cancer.
The main problem is that current chemotherapy drugs cannot be easily targeted. The drug particles flow in the bloodstream, damaging both healthy and cancerous cells. Typically, these drugs are flushed away quickly in organs such as the lungs and liver, limiting their effectiveness.
The remaining drugs are also unable to penetrate deep into the core of the tumor, leaving some cancer cells alive, which could lead to a resurgence in tumor growth.
Delivering anticancer drugs deep into tumors
Schematic of the apparatus used to investigate magnetic microbubble oscillation and nanoparticle release (credit: Yu Gao et al./NPG Asia Materials)
http://www.kurzweilai.net/images/magnetic-microbubble-nanoparticle-release-setup.jpg
The microbubbles are magnetic, so after injecting them into the bloodstream, they can be clustered around the tumor using magnets to ensure that they don’t kill the healthy cells, explains Xu, who has been working on cancer diagnosis and drug delivery systems since 2004.
“More importantly, our invention is the first of its kind that allows drug particles to be directed deep into a tumor in a few milliseconds. They can penetrate a depth of 50 cell layers or more (about 200 micrometers) — twice the width of a human hair. This helps to ensure that the drugs can reach the cancer cells on the surface and also inside the core of the tumor.”
According to Clinical Associate Professor Chia Sing Joo, a Senior Consultant at the Tan Tock Seng Hospital’s Endoscopy Centre and the Urology & Continence Clinic, “For anticancer drugs to achieve their best effectiveness, they need to penetrate into the tumor efficiently in order to reach the cytoplasm of all the cancer cells that are being targeted without affecting the normal cells.
“Currently, this can [only] be achieved by means of a direct injection into the tumor or by administering a large dosage of anticancer drugs, which can be painful, expensive, impractical and might have various side effects. If successful, I envisage [the new drug-delivery system] can be a good alternative treatment in the future, one which is low cost and yet effective for the treatment of cancers involving solid tumors, as it might minimize the side effects of drugs.” Joo is a surgeon experienced in the treatment of prostate, bladder and kidney cancer and a consultant for this study.
According to Ohl, an expert in biophysics who has published previous studies involving drug delivery systems and bubble dynamics, “most prototype drug delivery systems on the market face three main challenges before they can be commercially successful: they have to be non-invasive, patient-friendly, and yet cost-effective. We were able to come up with our solution that addresses these three challenges.”
The 12-person study team included scientists from City University of Hong Kong and Technion – Israel Institute of Technology (Technion). The team plans to use this new drug delivery system in studies on lung and liver cancer using animal models, and eventually clinical studies.
They estimate that it will take another eight to ten years before it reaches human clinical trials.
Abstract of Controlled nanoparticle release from stable magnetic microbubble oscillations
Magnetic microbubbles (MMBs) are microbubbles (MBs) coated with magnetic nanoparticles (NPs). MMBs not only maintain the acoustic properties of MBs, but also serve as an important contrast agent for magnetic resonance imaging. Such dual-modality functionality makes MMBs particularly useful for a wide range of biomedical applications, such as localized drug/gene delivery. This article reports the ability of MMBs to release their particle cargo on demand under stable oscillation. When stimulated by ultrasound at resonant frequencies, MMBs of 450 nm to 200 μm oscillate in volume and surface modes. Above an oscillation threshold, NPs are released from the MMB shell and can travel hundreds of micrometers from the surface of the bubble. The migration of NPs from MMBs can be described with a force balance model. With this technology, we deliver doxorubicin-containing poly(lactic-co-glycolic acid) particles across a physiological barrier bothin vitro and in vivo, with a 18-fold and 5-fold increase in NP delivery to the heart tissue of zebrafish and tumor tissue of mouse, respectively. The penetration of released NPs in tissues is also improved. The ability to remotely control the release of NPs from MMBs suggests opportunities for targeted drug delivery through/into tissues that are not easily diffused through or penetrated.
Artificial protein controls first self-assembly of C60 fullerenes

A Dartmouth College scientist and his collaborators* have created the first high-resolution co-assembly between a protein and buckminsterfullerene (C60), aka fullerene and buckyball (a sphere-like molecule composed of 60 carbon atoms and shaped like a soccer ball).
“This is a proof-of-principle study demonstrating that proteins can be used as effective vehicles for organizing nanomaterials by design,” says senior author Gevorg Grigoryan, an assistant professor of computer science at Dartmouth and senior author of a study discussed in an open-access paper in the journal in Nature Communications.
Proteins organize and orchestrate essentially all molecular processes in our cells. The goal of the new study was to create a new artificial protein that can direct the self-assembly of fullerene into ordered superstructures.

COP, a stable tetramer (a polymer derived from four identical single molecule) in isolation, interacts with C60 (fullerene) molecules via a surface-binding site and further self-assembles into a co-crystalline array called C60Sol–COP (credit: Kook-Han Kim et al./Nature Communications) http://www.kurzweilai.net/images/self-assembly-with-fullernene.jpg
Grigoryan and his colleagues show that that their artificial protein organizes a fullerene into a lattice called C60Sol–COP. COP, a protein that is a stable tetramer (a polymer derived from four identical single molecules), interacted with fullerene molecules via a surface-binding site and further self-assembled into an ordered crystalline superstructure. Interestingly, the superstructure exhibits high charge conductance, whereas both the protein-alone crystal and amorphous C60 are electrically insulating.
Grigoryan says that if we learn to do the programmable self-assembly of precisely organized molecular building blocks more generally, it will lead to a range of new materials with properties such as higher strength, lighter weight, and greater chemical reactivity, resulting in a host of applications, from medicine to energy and electronics.
Fullerenes are currently used in nanotechnology because of their high heat resistance and electrical superconductivity (when doped), but the molecule has been difficult to organize in useful ways.
* The study also included researchers from Dartmouth College, Sungkyunkwan University, the New Jersey Institute of Technology, the National Institute of Science Education and Research, the University of California-San Francisco, the University of Pennsylvania, and the Institute for Basic Science.
Abstract of Protein-directed self-assembly of a fullerene crystal
Learning to engineer self-assembly would enable the precise organization of molecules by design to create matter with tailored properties. Here we demonstrate that proteins can direct the self-assembly of buckminsterfullerene (C60) into ordered superstructures. A previously engineered tetrameric helical bundle binds C60 in solution, rendering it water soluble. Two tetramers associate with one C60, promoting further organization revealed in a 1.67-Å crystal structure. Fullerene groups occupy periodic lattice sites, sandwiched between two Tyr residues from adjacent tetramers. Strikingly, the assembly exhibits high charge conductance, whereas both the protein-alone crystal and amorphous C60 are electrically insulating. The affinity of C60 for its crystal-binding site is estimated to be in the nanomolar range, with lattices of known protein crystals geometrically compatible with incorporating the motif. Taken together, these findings suggest a new means of organizing fullerene molecules into a rich variety of lattices to generate new properties by design.
references:
- Kook-Han Kim, Dong-Kyun Ko, Yong-Tae Kim, Nam Hyeong Kim, Jaydeep Paul, Shao-Qing Zhang, Christopher B. Murray, Rudresh Acharya, William F. DeGrado, Yong Ho Kim, Gevorg Grigoryan. Protein-directed self-assembly of a fullerene crystal. Nature Communications, 2016; 7: 11429 DOI: 10.1038/NCOMMS11429 (open access)
- Supplementary Information
Protein-directed self-assembly of a fullerene crystal
Kook-Han Kim, Dong-Kyun Ko, Yong-Tae Kim, Nam Hyeong Kim,….., Yong Ho Kim & Gevorg Grigoryan
Nature Communications 2016;7,(11429) http://www.nature.com/ncomms/2016/160426/ncomms11429/full/ncomms11429.html
Programmable self-assembly of molecular building blocks is a highly desirable way of achieving bottom-up control over novel functions and materials. Applications of molecular assemblies are well explored in the literature, ranging from optoelectronic1, 2, magnetic3, and photovoltaic4 devices to chemical and bioanalytical sensing5, and medicine6. However, it has been a daunting challenge to quantitatively describe and control the driving forces that govern self-assembly, particularly given the broad range of molecular building blocks one would like to organize. In this respect, nature’s self-assembling macromolecules hold considerable promise as standard chassis for encoding precise organization. By learning to engineer the assembly of these molecules, myriad other molecular building blocks can be co-organized in desired ways through non-covalent or covalent attachment. The protein polymer is a particularly attractive candidate for a standard assembly chassis given its rich chemical alphabet, diversity of available assembly geometries, broad ability to engage other molecular moieties, and the possibility of engineered function. Considerable progress has been made in the area of engineering protein assemblies7, 8, using either computational9, 10,11, 12, 13, 14 or rational approaches15, 16, 17, 18, but the problem remains a grand challenge. A major difficulty lies in accounting for the enormous continuum of possible assembly geometries available to proteins to engineer a sequence that predictably prefers just one. General design principles, which provide predictive rules of assembly, are thus of enormous utility in limiting the geometric search space and enabling robust design11, 19.
In this work, we demonstrate the first ever high-resolution structure of co-assembly between a protein and buckminsterfullerene (C60), which suggests a simple structural mode for protein–fullerene co-organization. Three separate crystal structures, resolved to 1.67, 1.76 and 2.35 Å, reveal a protein lattice with C60 groups occupying periodic sites wedged between two helical segments, each donating a Tyr residue. A half site of the motif is estimated to have nM-scale affinity for C60, such that binding of fullerene appears to direct the organization of protein units in the co-crystal. The assembly exhibits a nm-spaced helical arrangement of fullerenes along a crystallographic axis, endowing the crystal with electrical conductance properties. We closely investigate the interfacial geometry of the C60-binding motif, finding it to be common among protein crystal lattices. C60 and its derivatives have been previously reported to interact with several proteins20, 21, 22, 23, 24, 25, although a high-resolution structure of a protein–C60 has been lacking. Still, prior evidence of interaction indicates that fullerenes and proteins are compatible as materials. This, together with the simple (and naturally recurrent) geometry of the C60-binding motif we discover, suggests that it may be possible to use the structural principles emergent from our study to generate a variety of C60–protein co-assemblies to further explore and exploit the properties of fullerenes26.
The aim of programmable self-assembly is to anticipate and harness unique collective properties that arise from precisely organized molecular building blocks. To this end, achieving atomic-level precision is crucial. This work demonstrates the first atomic resolution structures of a fullerene–protein assembly, establishing the feasibility of creating such objects, and further suggests a possible design principle for engineering such assemblies in general. How robust the discovered C60-binding motif is towards designing novel assemblies will need to be tested through a number of future design studies. However, the straightforward manner in which self-organization arose in our case, the simplicity of the C60-organizing motif in the lattice, together with its high affinity and the ubiquity of associated interfaces in natural protein lattices, are certainly promising with respect to the general applicability of the design principle. Our work also demonstrates the potential utility of exploring C60/protein co-organization, as derived supercrystals already showed synergistic charge conductance properties. Taken together, these results point to an exciting direction of inquiry towards generating protein–fullerene assemblies for the study and design of novel properties.
Scientists turn skin cells into heart and brain cells using only drugs — no stem cells required

Scientists at the Gladstone Institutes have used chemicals to transform skin cells into heart cells and brain cells, instead of adding external genes — making this accomplishment a breakthrough, according to the scientists.
The research lays the groundwork for one day being able to regenerate lost or damaged cells directly with pharmaceutical drugs — a more efficient and reliable method to reprogram cells and one that avoids medical concerns surrounding genetic engineering.
Instead, in two studies published in an open-access paper in Science and in Cell Stem Cell, the team of scientists at the Roddenberry Center for Stem Cell Biology and Medicine at Gladstone used chemical cocktails to gradually coax skin cells to change into organ-specific stem-cell-like cells and ultimately into heart or brain cells.
“This method brings us closer to being able to generate new cells at the site of injury in patients,” said Gladstone senior investigator Sheng Ding, PhD, the senior author on both studies. “Our hope is to one day treat diseases like heart failure or Parkinson’s disease with drugs that help the heart and brain regenerate damaged areas from their own existing tissue cells. This process is much closer to the natural regeneration that happens in animals like newts and salamanders, which has long fascinated us.”
Chemically Repaired Hearts
A human heart cell that was chemically reprogrammed from a human skin cell (credit: Nan Cao/Gladstone Institutes) http://www.kurzweilai.net/images/chemically-programmed-heart-cell.jpg
Transplanted adult heart cells do not survive or integrate properly into the heart and few stem cells can be coaxed into becoming heart cells.
Instead, in the Science study, the researchers used a cocktail of nine chemicals to change human skin cells into beating heart cells. By trial and error, they found the best combination of chemicals to begin the process by changing the cells into a state resembling multipotent stem cells (cells that can turn into many different types of cells in a particular organ). A second cocktail of chemicals and growth factors then helped transition the cells to become heart muscle cells.
With this method, more than 97% of the cells began beating, a characteristic of fully developed, healthy heart cells. The cells also responded appropriately to hormones, and molecularly, they resembled heart muscle cells, not skin cells. What’s more, when the cells were transplanted into a mouse heart early in the process, they developed into healthy-looking heart muscle cells within the organ.
“The ultimate goal in treating heart failure is a robust, reliable way for the heart to create new muscle cells,” said Srivastava, co-senior author on the Science paper. “Reprogramming a patient’s own cells could provide the safest and most efficient way to regenerate dying or diseased heart muscle.”
Rejuvenating the brain with neural stem cell-like cells
In the second study, authored by Gladstone postdoctoral scholar Mingliang Zhang, PhD, and published in Cell Stem Cell, the scientists created neural stem-cell-like cells from mouse skin cells using a similar approach.
The chemical cocktail again consisted of nine molecules, some of which overlapped with those used in the first study. Over ten days, the cocktail changed the identity of the cells, until all of the skin-cell genes were turned off and the genes of the neural stem-cell-like cells were gradually turned on.
When transplanted into mice, the neural stem-cell-like cells spontaneously developed into the three basic types of brain cells: neurons, oligodendrocytes, and astrocytes. The neural stem-cell-like cells were also able to self-replicate, making them ideal for treating neurodegenerative diseases or brain injury.
With their improved safety, these neural stem-cell-like cells could one day be used for cell replacement therapy in neurodegenerative diseases like Parkinson’s disease and Alzheimer’s disease, according to co-senior author Yadong Huang, MD, PhD, a senior investigator at Gladstone. “In the future, we could even imagine treating patients with a drug cocktail that acts on the brain or spinal cord, rejuvenating cells in the brain in real time.”
Gladstone Institutes | Chemically Reprogrammed Beating Heart Cell
Abstract of Conversion of human fibroblasts into functional cardiomyocytes by small molecules
Reprogramming somatic fibroblasts into alternative lineages would provide a promising source of cells for regenerative therapy. However, transdifferentiating human cells to specific homogeneous, functional cell types is challenging. Here we show that cardiomyocyte-like cells can be generated by treating human fibroblasts with a combination of nine compounds (9C). The chemically induced cardiomyocyte-like cells (ciCMs) uniformly contracted and resembled human cardiomyocytes in their transcriptome, epigenetic, and electrophysiological properties. 9C treatment of human fibroblasts resulted in a more open-chromatin conformation at key heart developmental genes, enabling their promoters/enhancers to bind effectors of major cardiogenic signals. When transplanted into infarcted mouse hearts, 9C-treated fibroblasts were efficiently converted to ciCMs. This pharmacological approach for lineage-specific reprogramming may have many important therapeutic implications after further optimization to generate mature cardiac cells.
Abstract of Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation
Cellular reprogramming using chemically defined conditions, without genetic manipulation, is a promising approach for generating clinically relevant cell types for regenerative medicine and drug discovery. However, small-molecule approaches for inducing lineage-specific stem cells from somatic cells across lineage boundaries have been challenging. Here, we report highly efficient reprogramming of mouse fibroblasts into induced neural stem cell-like cells (ciNSLCs) using a cocktail of nine components (M9). The resulting ciNSLCs closely resemble primary neural stem cells molecularly and functionally. Transcriptome analysis revealed that M9 induces a gradual and specific conversion of fibroblasts toward a neural fate. During reprogramming specific transcription factors such as Elk1 and Gli2 that are downstream of M9-induced signaling pathways bind and activate endogenous master neural genes to specify neural identity. Our study provides an effective chemical approach for generating neural stem cells from mouse fibroblasts and reveals mechanistic insights into underlying reprogramming processes.
references:
- Nan Cao, Yu Huang, Jiashun Zheng, C. Ian Spencer, Yu Zhang, Ji-Dong Fu, Baoming Nie, Min Xie, Mingliang Zhang, Haixia Wang, Tianhua Ma, Tao Xu, Guilai Shi, Deepak Srivastava, Sheng Ding. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science, 2016 DOI: 10.1126/science.aaf1502 (open access)
- Mingliang Zhang , Yuan-Hung Lin , Yujiao Jennifer Sun , Saiyong Zhu10 , Jiashun Zheng , Kai Liu , Nan Cao , Ke Li , Yadong Huang , Sheng Ding. Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell, 2016 DOI: 10.1016/j.stem.2016.03.020
Ultrafast laser technique identifies brain tumors in real time
A research group at VU University Amsterdam (The Netherlands) has shown that an ultrafast laser technique can reveal exactly where brain tumors are, producing images in less than a minute and enabling surgeons to removetumors without compromising healthy tissue.
Related: OCT-based approach facilitates brain cancer surgery
Pathologists typically use staining methods, in which chemicals like hematoxylin and eosin turn different tissue components blue and red, revealing its structure and whether there are any tumor cells. But for a definitive diagnosis, this process can take up to 24 hours—which means surgeons may not realize some cancerous tissue has escaped from their attention until after surgery, requiring a second operation and more risk.
But the research team’s new ultrafast laser technique is label-free—instead, they fire short, 20-fs-long laser pulses into the tissue, and when three photons converge at the same time and place, the photons interact with the nonlinear optical properties of the tissue. Through well-known phenomena in optics called second- and third-harmonic generation, these interactions produce a single photon.
The key is that the incoming and outgoing photons have different wavelengths. The incoming photons are at 1200 nm, long enough to penetrate deep into the tissue. The single photon that is produced, however, is at 600 or 400 nm, depending on if it’s second- or third-harmonic generation. The shorter wavelengths mean the photon can scatter in the tissue. The scattered photon thus contains information about the tissue, and when it reaches a detector—in this case, a high-sensitivity gallium arsenide phosphide (GaAsP) photomultiplier tube—it reveals what the tissue looks like inside.

Tissue from a patient diagnosed with low-grade glioma. The green image is taken with the new method, while the pink uses conventional hematoxylin and eosin staining. Going from the upper left to the lower right, both images show increasing cell density because of more tumor tissue. The insets reveal the high density of tumor cells. (Credit: N.V. Kuzmin et al, VU University Amsterdam, The Netherlands)
The research team used the technique to analyze glial brain tumors, which are particularly deadly because it’s hard to get rid of tumor cells by surgery, irradiation, and chemotherapy without substantial collateral damage to the surrounding brain tissue. They tested their method on samples of glial brain tumors from humans, finding that the histological detail in these images was as good—if not better—than those made with conventional staining techniques. They were able to make most images in under a minute. The smaller ones took less than a second, while larger images of a few square millimeters took five minutes—making it possible to do it in real time in the operating room, according to Marloes Groot of VU University Amsterdam, who led the work.
Now that they’ve shown their approach works, the researchers are developing a handheld device that a surgeon can use to identify a tumor’s border during surgery. The incoming laser pulses can only reach a depth of about 100 µm into the tissue. To reach farther, Groot envisions attaching a needle that can pierce the tissue and deliver photons deeper, allowing diagnosis during an operation and possibly before surgery begins.
Full details of the work appear in the journal Biomedical Optics Express; for more information, please visit http://dx.doi.org/10.1364/boe.7.001889.
Third harmonic generation imaging for fast, label-free pathology of human brain tumors
N. V. Kuzmin, P. Wesseling, P. C. de Witt Hamer, D. P. Noske, G. D. Galgano, H. D. Mansvelder, J. C. Baayen, and M. L. Groot
Biomedical Optics Express > Volume 7 > Issue 5 > Page 1889
In brain tumor surgery, recognition of tumor boundaries is key. However, intraoperative assessment of tumor boundaries by the neurosurgeon is difficult. Therefore, there is an urgent need for tools that provide the neurosurgeon with pathological information during the operation. We show that third harmonic generation (THG) microscopy provides label-free, real-time images of histopathological quality; increased cellularity, nuclear pleomorphism, and rarefaction of neuropil in fresh, unstained human brain tissue could be clearly recognized. We further demonstrate THG images taken with a GRIN objective, as a step toward in situ THG microendoscopy of tumor boundaries. THG imaging is thus a promising tool for optical biopsies.

Leave a Reply