Evolution of Myoglobin and Hemoglobin
Author and Curator: Larry H. Bernstein, MD, FCAP
Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases
Paul R. Gardner
Journal of Inorganic Biochemistry Jan 2005; 99(1): 247–266
http://dx.doi.org:/10.1016/j.jinorgbio.2004.10.003
Microbial flavohemoglobins (flavoHbs) and hemoglobins (Hbs) show large radical dotNO dioxygenation rate constants ranging from 745 to 2900 μM−1 s−1 suggesting a primal radical dotNO dioxygenase (NOD) (EC 1.14.12.17) function for the ancient Hb superfamily. Indeed, modern O2-transporting and storing mammalian red blood cell Hb and related muscle myoglobin (Mb) show vestigial radical dotNO dioxygenation activity with rate constants of 34–89 μM−1 s−1. In support of a NOD function, microbial flavoHbs and Hbs catalyze O2-dependent cellular radical dotNO metabolism, protect cells from radical dotNO poisoning, and are induced by radical dotNO exposures. Red blood cell Hb, myocyte Mb, and flavoHb-like activities metabolize radical dotNO in the vascular lumen, muscle, and other mammalian cells, respectively, decreasing radical dotNO signalling and toxicity. HbFe(III)–OOradical dot, HbFe(III)–OONO and protein-caged [HbFe(III)–Oradical dotradical dotNO2] are proposed intermediates in a reaction mechanism that combines both O-atoms of O2 with radical dotNO to form nitrate and HbFe(III). A conserved Hb heme pocket structure facilitates the dioxygenation reaction and efficient turnover is achieved through the univalent reduction of HbFe(III) by associated reductases. High affinity flavoHb and Hb heme ligands, and other inhibitors, may find application as antibiotics and antitumor agents that enhance the toxicity of immune cell-derived radical dotNO or as vasorelaxants that increase radical dotNO signaling.
http://ars.els-cdn.com/content/image/1-s2.0-S016201340400296X-gr1.sml
http://ars.els-cdn.com/content/image/1-s2.0-S016201340400296X-gr2.sml
http://ars.els-cdn.com/content/image/1-s2.0-S016201340400296X-gr3.sml
The evolution of the globin family genes: Concordance of stochastic and augmented maximum parsimony genetic distances for α hemoglobin, β hemoglobin and myoglobin phylogenies
R Holmquist, TH Jukes, H Moise, M Goodman, GW Moore
Journal of Molecular Biology Jul 1976; 105(1): 39–74
http://dx.doi.org:/10.1016/0022-2836(76)90194-7
We compare the amino acid sequences of 70 globing, representing the following families: (a) α hemoglobin chains; (b) β hemoglobin chains; (c) myoglobins; (d) two lamprey, a mollusc, and two plant globins. The comparisons show a convergence of maximal and minimal estimates of genetic differences as calculated respectively by the stochastic and maximum parsimony procedures, thus demonstrating for the first time the logical consistency and complementarity of the two procedures. Evolutionary rates are non-constant, varying over a range of 1 to 75 nucleotide replacements per 100 codons per 108 years. These rate differentials are resolved into two components (a) due to change in the number of codon sites free to fix mutations during the period of divergence of the species involved; (b) due to change in fixation intensity at each site. These two components also show non-uniformity along different lineages. Positive Darwinian natural selection can bring about an increase in either component, and negative or stabilizing selection in protein evolution can lead to decreases. Accelerated rates of globin evolution were found in lineages of cold-blooded vertebrates, some marsupials, and early placental mammals, while slower rates were found in warm-blooded vertebrates, especially higher primates. One manifestation of negative selection in the globins is that minimal 3-base type amino acid replacements occur less frequently than would be expected if base replacements had occurred and were accepted at random. The selection against these replacements is not due to atypical behavior with respect to the change in electrical charge involved in the replacements. Interestingly, the globins from the lamprey, sea hare and the legumes are as distant from one another as are α-hemoglobin and β-hemoglobin from myoglobin.
Hemoglobin Orthologs
http://www.bio.davidson.edu/Courses/Molbio/MolStudents/spring2005/Heiner/ortholog.html
Orthologs are sequences of genes that evolved from a common ancestor and can be traced evolutionarily through different species. By comparing the ortholog sequences of a specific gene between many species, the amino acid sequences which are conserved can be determined. These highly conserved sequences are important, because they provide information on which amino acids are essential to the protein structure and function.
Evolution of Hemoglobin
Hemoglobin is derived from the myoglobin protein, and ancestral species just had myoglobin for oxygen transport. 500 million years ago the myoglobin gene duplicated and part of the gene became hemoglobin. Lampreys are the most ancestral animal to have hemoglobin, and the ancestral version was composed of dimers instead of tetramers and was only weakly cooperative. 100 million years later, the hemoglobin gene duplicated again forming alpha and beta subunits. This form of derived hemoglobin is found in bony fish, reptiles, and mammals, which all have both alpha and beta subunits to form a tetramer (Mathews et al., 2000).
Conserved Sequences
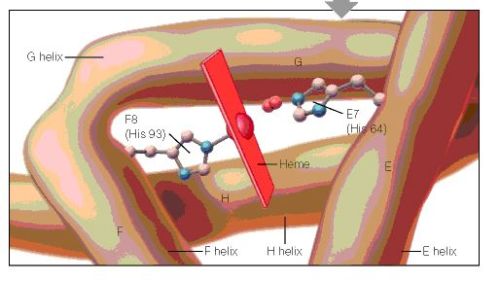
When the amino acid sequences of myoglobin, the hemoglobin alpha subunit, and the hemoglobin beta subunit are compared, there are several amino acids that remain conserved between all three globins (Mathews et al., 2000). These amino acid sequences are considered truly essential, because they have remained unchanged throughout evolution, and therefore are fundamental to the function of the protein. These essential amino acids can be seen in Figure 1, which compares myoglobin, and the alpha and beta subunits of hemoglobin. The histidines in helix F8 and helix E7 are highly conserved. These histidines are located proximally and distally to the heme molecule and keep the heme molecule in place within the hemoglobin protein as seen in Figure 2 (Mathews et al., 2000). This shows that the position of the heme molecule within the globin protein is essential to its function. Likewise, the amino acids in the FG region are also highly conserved. This region of the protein is essential to the conformational change between the T to R states (Mathews et al., 2000). Additionally, the amino acids at the alpha-beta subunit interfaces are highly conserved, because they also affect the conformational change between the subunits, which regulates oxygen affinity and cooperativity. In general, the most highly conserved sequences are located within the interior of the hemoglobin protein where the subunits contact each other (Gribaldo et al., 2003).
http://www.bio.davidson.edu/Courses/Molbio/MolStudents/spring2005/Heiner/figure2.jpg
Figure 2: A cartoon drawing of the structure of hemoglobin around heme molecule. The histadines in helix F8 and E7 interact directly with the heme molecule. http://www.aw-bc.com/mathews/ch07/fi7p5.htm (permission pending).
Figure 1: The amino acid sequences of myoglobin, alpha subunit of hemoglobin, and beta subunit of hemoglobin. The amino acid sequences highlighted in tan are conserved between all three globins and the amino acid sequences highlighted in gray are conserved between alpha and beta hemoglobin. http://www.aw-bc.com/mathews/ch07/fi7p11.htm (permission pending).
http://www.bio.davidson.edu/Courses/Molbio/MolStudents/spring2005/Heiner/figure1.jpg
Figure 2: A cartoon drawing of the structure of hemoglobin around heme molecule. The histidines in helix F8 and E7 interact directly with the heme molecule. http://www.aw-bc.com/mathews/ch07/fi7p11.htm (permission pending).
Alpha Subunit of Hemoglobin
The alpha subunit of hemoglobin has several amino acid sequences that are conserved across many species and are essential to its function. The alpha subunit of hemoglobin is encoded by the 2 genes HBA1 and HBA2 both located on chromosome 16 (GeneCard, 2005). Click here to see the gene card for HBA1. To determine which amino acid sequences are conserved, I compared the orthologs of HBA1 in Homo sapiens (humans) to 5 additional species including, Xenopus tropicalis (African clawed frog), Danio rerio (Zebra fish), Gallus gallus (Red jungle fowl), Mus musculus (mouse), and Rattus norvegicus (rat) using the Ensembl program. Figure 3 shows the 6 orthologs aligned and the important conserved regions highlighted. The stars indicate amino acids that are conserved between all of the species. As a general observation, the mouse ortholog of HBA is the most similar to human HBA, because it is the most evolutionarily related. The amino acid sequences that are conserved in all globin proteins (highlighted in blue) can be seen in Figure 3. There are also several conserved amino acids that are specifically important to HBA structure (highlighted in red) including: the phenylalanine (F) at position 44, which is in direct contact with the heme group; tyrosine (Y) at position 142, which stabilizes the hemoglobin molecule by forming hydrogen bonds between two of the helices; and glycine (G) at position 26, which is small and therefore allows two of the helices to approach each other, which is important to the structure of hemoglobin (Natzke, 1998). Additionally, there are several proteins found in the alpha subunit that are involved in the movement of the alpha and beta subunits (also highlighted in red) including: the tyrosine (Y) at position 43, which interacts with the beta subunit during the R state, and the arginine (N) at position 143, which interacts with the beta subunit during the T state (Gribaldo et al., 2003).
Mutations
Looking at the effects mutated portions of a gene is also a good way to determine the function of highly conserved sequences. In hemoglobin, deleterious mutations are most common in the heme pockets of the protein and in the alpha and beta subunit interfaces (Mathews et al., 2000). There are several key mutations in highly conserved portions of HBA (highlighted in yellow) including: the substitution of histidine (H) at position 88 to tyrosine (Y), which disrupts the heme molecule leading to decreased oxygen affinity; the substitution of arginine (N) at position 143 to histidine (H), which eliminates a bond in the T state and therefore favors the R state, resulting in increased oxygen affinity; the substitution of proline (P) at position 97 to arginine (N), which alters the alpha-beta contact region and results in the disassociation of the hemoglobin complex; and the substitution of leucine (L) at position 138 for proline (P), which interrupts the helix formation and also results in the disassociation of the hemoglobin complex (Mathews et al., 2000).
Bar-headed Goose Hemoglobin
As mentioned on the previous page, the bar-headed goose has hemoglobin that is specifically adapted to high altitudes. The bar-headed goose hemoglobin has an increased oxygen affinity which allows it to live in low oxygen pressure environments (Liang et al., 2001). This increased oxygen affinity is the result of a mutation at position 121 in the alpha subunit, which is highly conserved in other species, from proline to alanine, as seen in Figure 4 (Liang et al., 2001). This substitution leaves a two-carbon gap between the alpha-beta dimer, which relaxes the T structure and allows it to bind oxygen more readily under lower pressures (Jessen et al. 1991). Thus, comparing orthologs can also be used to explain differences in the oxygen binding capabilities of hemoglobin in different species.
References
Ensembl. Ensembl Genome Browser. http://www.ensembl.org/. Accessed March 2005.
GeneCard. 2005. GeneCard for HBA1. http://genome-www.stanford.edu/cgi-bin/genecards/carddisp?HBA1&search=HBA&suff=txt. Accessed March 2005.
Gribaldo, Simonetta, Didier Casane, Philippe Lopez and Herve Philippe. 2003. Functional Divergence Prediction from Evolutionary Analysis: A Case Study of Vertebrate Hemoglobin. Molecular Biology and Evolution 20 (11): 1754-1759.
Jessen, Timm H et al. 1991. Adaptation of bird hemoglobins to high altitudes: Demonstration of molecular mechanism by protein engineering. Evolution 88: 6519-6522.
Liang, Yuhe et al. 2001. The Crystal Structure of Bar-headed Goose Hemoglobin in Deoxy Form: The Alloseteric Mechanism of a Hemoglobin Species with High Oxygen Affinity. Journal of Molecular Biology 313: 123-137.
Mathews, Christopher, Kensal Van Holde and Kevin Ahern. 2000. Biochemistry 3 rd edition. http://www.aw-bc.com/mathews/ch07/c07emhp.htm . Accessed March 2005.
Natzke, Lisa. 1998. Hemoglobin. http://biology.kenyon.edu/BMB/Chime/Lisa/FRAMES/hemetext.htm. Accessed March 2005.
Divergence pattern and selective mode in protein evolution: the example of vertebrate myoglobins and hemoglobin chains.
Otsuka J1, Miyazaki K, Horimoto K.
J Mol Evol. 1993 Feb; 36(2):153-81.
The evolutionary relation of vertebrate myoglobin and the hemoglobin chains including the agnathan hemoglobin chain is investigated on the basis of a new view of amino acid changes that is developed by canonical discriminant analysis of amino acid residues at individual sites. In contrast to the clear discrimination of amino acid residues between myoglobin, hemoglobin alpha chain, and hemoglobin beta chain in warm-blood vertebrates, the three types of globins in the lower class of vertebrates show so much variation that they are not well discriminated. This is seen particularly at the sites that are ascertained in mammals to carry the amino acid residues participating in stabilizing the monomeric structure in myoglobin and the residues forming the subunit contacts in hemoglobin. At these sites, agnathan hemoglobin chains are evaluated to be intermediate between the myoglobin and hemoglobin chains of gnathostomes. The variation in the phylogenetically lower class of globins is also seen in the internal region; there the amino acid residues of myoglobin and hemoglobin chains in the phylogenetically higher class exhibit an example of parallel evolution at the molecular level. New quantities, the distance of sequence property between discriminated groups and the variation within each group, are derived from the values of discriminant functions along the peptide chain, and this set of quantities simply describes an overall feature of globins such that the distinction between the three types of globins has been clearer as the vertebrates have evolved to become jawed, landed, and warm-blooded. This result strongly suggests that the functional constraint on the amino acid sequence of a protein is changed by living conditions and that severe conditions constitute a driving force that creates a distinctive protein from a less-constrained protein.
The globin gene repertoire of lampreys: Convergent evolution of hemoglobin and myoglobin in jawed and jawless vertebrates
K Schwarze, KL Campbell, T Hankeln, JF Storz, FG Hoffmann and T Burmester
Mol Biol Evol (2014). http://dx.doi.org:/10.1093/molbev/msu216
Agnathans (jawless vertebrates) occupy a key phylogenetic position for illuminating the evolution of vertebrate anatomy and physiology. Evaluation of the agnathan globin gene repertoire can thus aid efforts to reconstruct the origin and evolution of the globin genes of vertebrates, a superfamily that includes the well-known model proteins hemoglobin and myoglobin. Here we report a comprehensive analysis of the genome of the sea lamprey (Petromyzon marinus) which revealed 23 intact globin genes and two hemoglobin pseudogenes. Analyses of the genome of the Arctic lamprey (Lethenteron camtschaticum) identified 18 full length and five partial globin gene sequences. The majority of the globin genes in both lamprey species correspond to the known agnathan hemoglobins. Both genomes harbor two copies of globin X, an ancient globin gene that has a broad phylogenetic distribution in the animal kingdom. Surprisingly, we found no evidence for an ortholog of neuroglobin in the lamprey genomes. Expression and phylogenetic analyses identified an ortholog of cytoglobin in the lampreys; in fact, our results indicate that cytoglobin is the only orthologous vertebrate-specific globin that has been retained in both gnathostomes and agnathans. Notably, we also found two globins that are highly expressed in the heart of P. marinus, thus representing functional myoglobins. Both genes have orthologs in L. camtschaticum. Phylogenetic analyses indicate that these heart-expressed globins are not orthologous to the myoglobins of jawed vertebrates (Gnathostomata), but originated independently within the agnathans. The agnathan myoglobin and hemoglobin proteins form a monophyletic group to the exclusion of functionally analogous myoglobins and hemoglobins of gnathostomes, indicating that specialized respiratory proteins for O2 transport in the blood and O2 storage in the striated muscles evolved independently in both lineages. This dual convergence of O2-transport and O2-storage proteins in agnathans and gnathostomes involved the convergent co-option of different precursor proteins in the ancestral globin repertoire of vertebrates.
Globin evolution
Kent Holsinger
http://darwin.eeb.uconn.edu/eeb348/lecturenotes/molevol-multigene/node2.html
I’ve just pointed out the distinction between myoglobin and hemoglobin. You may also remember that hemoglobin is a multimeric protein consisting of four subunits, 2 α\alpha subunits and 2 β\beta subunits. What you may not know is that in humans there are actually two types of α\alpha hemoglobin and four types of β\beta hemoglobin, each coded by a different genetic locus (see Table 1). The five α\alpha -globin loci (α\alpha_1, α\alpha_2, ς\zeta, and two non-functional pseudogenes) are found in a cluster on chromosome 16. The six β\beta-globin loci (ε\epsilon, ϒ\gamma_G, ϒ\gamma_A, δ\delta, β\beta, and a pseudogene) are found in a cluster on chromosome 11. The myoglobin locus is on chromosome 22.
Table 1: Human hemoglobins arranged in developmental sequence. Adult hemoglobins composed of 2 and 2 subunits typically account for less than 3% of hemoglobins in adults (http://sickle.bwh.harvard.edu/hbsynthesis.html).
Not only do we have all of these different types of globin genes in our bodies, they’re all related to one another. Comparative sequence analysis has shown that vertebrate myoglobin and hemoglobins diverged from one another about 450 million years ago. Figure 1 shows a phylogenetic analysis of globin genes from humans, mice, and a variety of Archaea. Focus your attention on the part of the tree that has human and mouse sequences. You’ll notice two interesting things:
Human and mouse neuroglobins (Ngb) are more closely related to one another than they are to other globins, even those from the same species. The same holds true for cytoglobins (Cyg) and myoglobins (Mb).
Within the hemoglobins, only mouse β\beta-globin (Mouse HbB) is misplaced. All other α\alpha- and β\beta-globins group with the corresponding mouse and human loci.
This pattern is exactly what we expect as a result of duplication and divergence. Up to the time that a gene becomes duplicated, its evolutionary history matches the evolutionary history of the organisms containing it. Once there are duplicate copies, each follows an independent evolutionary history. Each traces the history of speciation and divergence. And over long periods duplicate copies of the same gene share more recent common ancestry with copies of the same gene in a different species than they do with duplicate genes in the same genome.
Figure 1: Evolution of globin genes in Archaea and mammals (from [2]).
http://darwin.eeb.uconn.edu/eeb348/lecturenotes/molevol-multigene/img11.png
A history of duplication and divergence in multigene families makes it important to distinguish between two classes of related loci: those that represent the same locus in different species and between which divergence is a result of species divergence are orthologs. Those that represent different loci and between which divergence occurred after duplication of an ancestral gene are paralogs. The β\beta-globin loci of humans and chickens are orthologous. The α\alpha $- and $\beta $-globin loci of any pair of taxa are paralogous.
As multigene families go, the globin family is relatively simple and easy to understand. There are only about a dozen loci involved, one isolated locus (myoglobin) and two clusters of loci ($\alpha- and β\beta-globins). You’ll find a diagram of the β\beta-globin cluster in Figure 2. As you can see the β\beta-globins are not only evolutionarily related to one another they occur relatively close to one another on chromosome 11 in humans.
Figure 2: Structure of the human β\beta-globin gene cluster. % identity refers to similarity to the mouse β\beta-globin sequence. From http://globin.cse.psu.edu/html/pip/betaglobin/iplot.ps (retrieved 28 Nov 2006).
Other families are far more complex. Class I and class II MHC loci, for example are part of the same multigene family. Moreover, immunoglobulins, T-cell receptors, and, and MHC loci are part of a larger superfamily of genes, i.e., all are ultimately derived from a common ancestral gene by duplication and divergence. Table 2 lists a few examples of multigene families and superfamilies in the human genome and the number of proteins produced.
| Table 2: A few gene families from the human genome (adapted from [5,6]). |
Distribution and conservation of sequence motifs throughout mammalian beta-globin gene clusters.A detailed map of the gene cluster is shown on the numbered line
the evolutionary history of three hypothetical living species (C, D, and E), inferred by comparing amino-acid differences in their myoglobin molecules.
http://media-1.web.britannica.com/eb-media/98/52998-004-A8682A5B.jpg
http://i.stack.imgur.com/WQJe9.jpg
myoglobin hs much higher oxygen affinity than Hb
Evolution of Myoglobin / Hemoglobin Proteins
Primitive Globin – Very primitive animals had only a myoglobin-like, single-chain ancestral globin for oxygen storage and were so small that they did not require a transport protein. Roughly 500 million years ago the ancestral myoglobin gene was duplicated. One copy became the ancestor of the myoglobin genes of all higher organisms. The other copy evolved into the gene for an oxygen transport protein and gave rise to the hemoglobins.
Most Primitive Hemoglobin – The most primitive animals to possess hemoglobin are the lampreys. Lamprey hemoglobin can form dimers but not tetramers and is only weakly cooperative. It represents a first step toward allosteric binding. Subsequently a second gene duplication must have occurred, giving rise to the ancestors of the present-day and hemoglobin chain families. This must have happened about 400 million years ago, at about the time of divergence of the sharks and bony fish. The evolutionary line of the bony fish led to the reptiles and eventually to the mammals, all carrying genes for both and globins and capable of forming tetrameric 22 hemoglobins. Further gene duplications have occurred in the hemoglobin line, leading to the embryonic forms and , the fetal form, , and the infant form (Figure 7.22).
Conserved Amino Acid Sequences – During the long evolution of the myoglobin/hemoglobin family of proteins, only a few amino acid residues have remained invariant (Figure 7.11). They include the histidines proximal and distal to the heme iron (F8 and E7- see Figure 7.5b) and Val FG5, which has been implicated in the hemoglobin deoxy/oxy conformation change. These may mark the truly essential positions in the molecule. Other regions highly conserved in hemoglobins are those near the 1 – 2 and 2 – 1 contacts. These parts of the molecule are most directly involved in the allosteric conformational change.