Protein regulator of HIV replication
Larry H. Bernstein, MD, FCAP, Curator
LPBI
Updated 11/26/2015
Closing the loop on an HIV escape mechanism
University of Delaware
http://www.rdmag.com/news/2015/11/closing-loop-hiv-escape-mechanism

http://www.rdmag.com/sites/rdmag.com/files/newsletter-ads/CHEM-Polenova_Research_Groups-111015-015.jpg
Tatyana Polenova, professor of chemistry and biochemistry at UD (background, left), with her UD research team involved in the HIV study. Next to her is Huilan Zhang. In the foreground, from left, are Guangjin Hou and Manman Lu.
Nearly 37 million people worldwide are living with HIV. When the virus destroys so many immune cells that the body can’t fight off infection, AIDS will develop. The disease took the lives of more than a million people last year.
For the past three and a half years, a team of researchers from six universities, led by the University of Delaware and funded by the National Institutes of Health and the National Science Foundation, has been working to uncover new information about a protein that regulates HIV’s capability to hijack a cell and start replicating. Their findings, reported recently in the Proceedings of the National Academy of Sciences, point to a new avenue for developing potential strategies to thwart the virus.
The team included scientists from UD, the University of Pittsburgh School of Medicine, University of Illinois at Urbana-Champaign, Carnegie Mellon University, the National High Magnetic Field Laboratory at Florida State University and Vanderbilt University School of Medicine. They used a combination of high-tech tools and techniques, including magic-angle-spinning nuclear magnetic resonance (NMR) spectroscopy and computer simulations of molecules, to examine the interactions between HIV and the host-cell protein cyclophilin A (CypA), right down to the movement of individual atoms.
“In a nutshell, we found that the infectivity of HIV is regulated by the motions of these proteins,” says Tatyana Polenova, professor of chemistry and biochemistry at the University of Delaware, who led the study. “It’s a subtle regulation strategy that does not involve major structural changes in the virus.”
Sixty times smaller than a red blood cell, HIV contains a cone-shaped shell, or capsid, made of protein, which surrounds two strands of RNA and the enzymes the virus needs for replication. Like any virus, HIV can only produce copies of itself once it has invaded a host organism. Then it will begin directing certain host cells to begin producing the virus.
But how does HIV invade a cell? In humans, the protein CypA can either promote or inhibit viral infection through interactions with the HIV capsid, although the exact mechanism is not yet known. A portion of the HIV capsid protein, called the CypA loop, is responsible for binding to the CypA in the human host cell. Once this occurs, the virus typically becomes infectious.
However, a change of just one amino acid in the CypA loop can cause the virus to operate opposite from how it does normally, allowing the virus to become non-infectious when CypA is present, and to become infectious when there is no CypA present. Such changes are called “escape mutations,” Polenova says, because they allow the virus to “escape” from its dependence on CypA.
To home in on this escape mechanism, the research team examined assemblies of different variants of HIV capsid protein complexed with CypA. Using magic-angle-spinning NMR, they recorded the motions in these assemblies, atom by atom, on time scales ranging from nanoseconds to milliseconds, from a billionth of a second to a thousandth of a second.
The team found that a reduction in the naturally occurring motions in the binding region due to the mutations allowed the virus to escape from CypA dependence. Magic-angle-spinning NMR experiments provided a direct probe of these motions, recording the changes in the magnetic interactions between nuclei. Computer simulations allowed the team to visualize the motions.
Some portions of the capsid protein do not move at all or move only a little while other portions undergo large-amplitude motions distributed over a wide range of time scales, with the most dynamic region being the CypA loop. Polenova says it is rather surprising that such extensive motions are present in the assembled capsid, and that these dynamics could be detected by both NMR and computer simulations.
“It is the first time that quantitative agreement between experiment and computation was achieved in a dynamics study, and it’s particularly exciting that this was attained for such a complex system,” Polenova says. “We hope this work may guide the development of new therapeutic interventions, such as small molecules that would serve as interactors with the HIV capsid and inhibit these dynamics.”
Polenova says the diverse team of researchers, with expertise in HIV virology, structural biology, biophysics and biochemistry, was critical to the study’s success, along with access to national high-field NMR facilities through the National High Magnetic Field Laboratory. The team was assembled through the NIH-funded Pittsburgh Center for HIV Protein Interactions. Led by Prof. Angela Gronenborn, the center brings together high-caliber scientists and facilities to elucidate the interactions of HIV proteins with host cell factors.
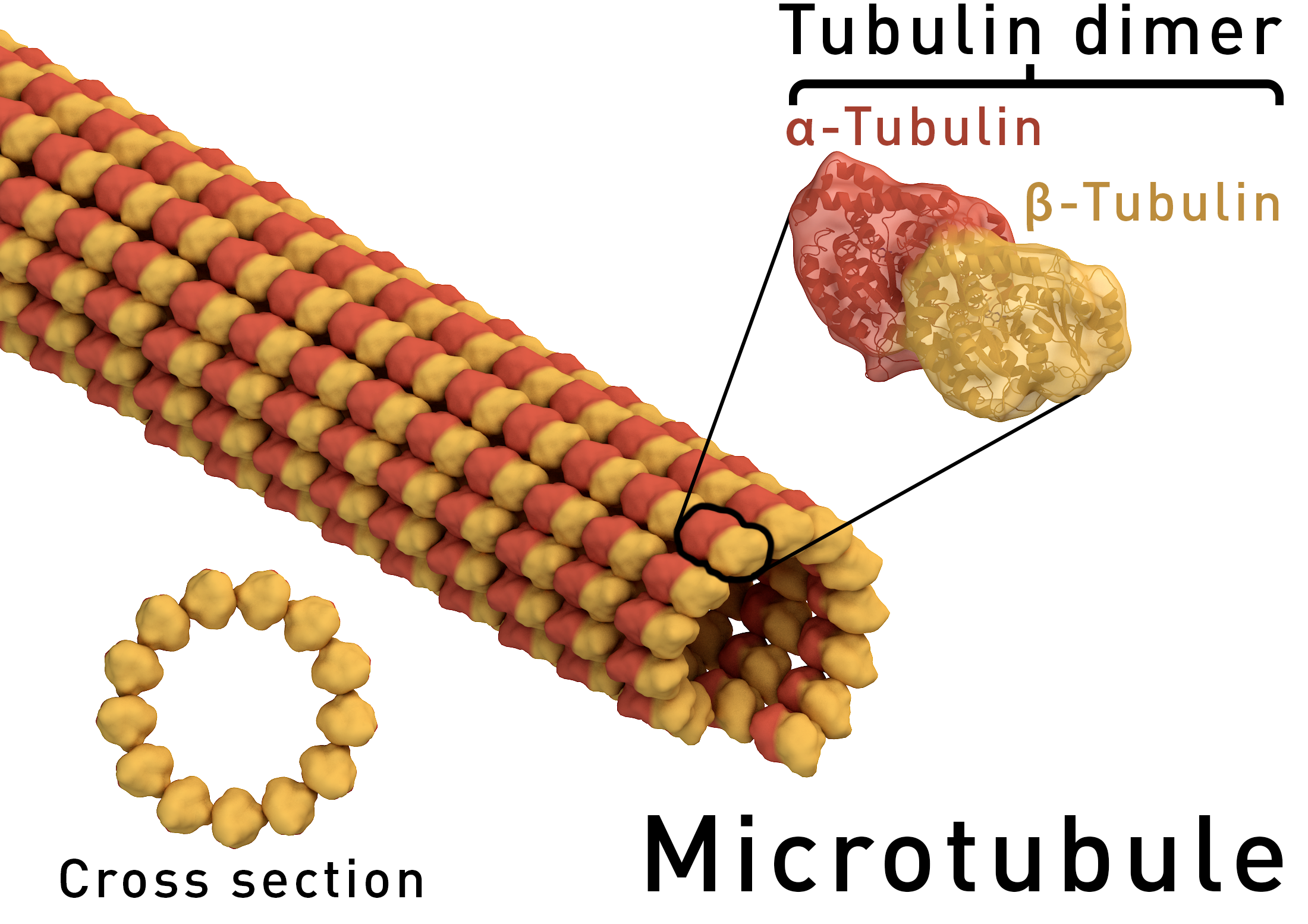
Atomic-resolution structure of the CAP-Gly domain of dynactin on polymeric microtubules determined by magic angle spinning NMR spectroscopy
Si Yana,1, Changmiao Guoa,1, Guangjin Houa, Huilan Zhanga, Xingyu Lua, John Charles Williamsb, and Tatyana Polenovaa,2 Author Affiliations
PNAS Nov 24, 2015; 112(47): 14611–14616 http://dx.doi.org:/10.1073/pnas.1509852112
Significance
Microtubules and their associated proteins are central to most cellular functions. They have been extensively studied at multiple levels of resolution; however, significant knowledge gaps remain. Structures of microtubule-associated proteins bound to microtubules are not known at atomic resolution. We used magic angle spinning NMR to solve a structure of dynactin’s cytoskeleton-associated protein glycine-rich (CAP-Gly) domain bound to microtubules and to determine the intermolecular interface, the first example, to our knowledge, of the atomic-resolution structure of a microtubule-associated protein on polymeric microtubules. The results reveal remarkable structural plasticity of CAP-Gly, which enables CAP-Gly’s binding to microtubules and other binding partners. This approach offers atomic-resolution information of microtubule-binding proteins on microtubules and opens up the possibility to study critical parameters such as protonation states, strain, and dynamics on multiple time scales.
Microtubules and their associated proteins perform a broad array of essential physiological functions, including mitosis, polarization and differentiation, cell migration, and vesicle and organelle transport. As such, they have been extensively studied at multiple levels of resolution (e.g., from structural biology to cell biology). Despite these efforts, there remain significant gaps in our knowledge concerning how microtubule-binding proteins bind to microtubules, how dynamics connect different conformational states, and how these interactions and dynamics affect cellular processes. Structures of microtubule-associated proteins assembled on polymeric microtubules are not known at atomic resolution. Here, we report a structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain of dynactin motor on polymeric microtubules, solved by magic angle spinning NMR spectroscopy. We present the intermolecular interface of CAP-Gly with microtubules, derived by recording direct dipolar contacts between CAP-Gly and tubulin using double rotational echo double resonance (dREDOR)-filtered experiments. Our results indicate that the structure adopted by CAP-Gly varies, particularly around its loop regions, permitting its interaction with multiple binding partners and with the microtubules. To our knowledge, this study reports the first atomic-resolution structure of a microtubule-associated protein on polymeric microtubules. Our approach lays the foundation for atomic-resolution structural analysis of other microtubule-associated motors.
How Viruses Commandeer Human Proteins
http://www.technologynetworks.com/Proteomics/news.aspx?ID=185156
Researchers have produced the first image of an important human protein as it binds with ribonucleic acid (RNA), a discovery that could offer clues to how some viruses, including HIV, control expression of their genetic material.
RNA is one of three macromolecules — along with DNA and proteins — essential to all forms of life. By understanding how hnRNP A1 binds to RNA, the scientists may find ways to jam up components of the replication machinery when the protein is coopted by disease.
The team of scientists reveals the mechanism used by the protein, hnRNP A1 to link to the section of RNA, called the ‘hairpin loop.’
They found that hnRNP A1, a protein essential to cell function and virus replication, has a significantly different structure than its only previously known form: binding to DNA.
“We solved the three-dimensional structure of the protein bound to an RNA hairpin derived from the HIV virus,” said Blanton Tolbert, a chemistry professor at Case Western Reserve. “But because the hairpin loop is found in other viruses and throughout healthy cells, our findings may help explain how the protein connects to the other hairpin targets.”
Tolbert began this research six years ago, frustrated that the only information available was the structure of the protein bound to a synthetic DNA, which isn’t its natural target.
Proteins that bind hairpins sense both the structure and the sequence information presented in the loop. The structure of the DNA complex did not demonstrate the molecular recognition that must take place to bind RNA hairpins.
The process
To discover the structure bound to RNA, the researchers combined three techniques: X-ray crystallography, nuclear magnetic resonance spectroscopy and small angle x-ray scattering. Each technique yielded a piece of the puzzle.
To bind to RNA, hnRNP A1 has two domains, RRM1 and RRM2, which are akin to hands. Scientists already knew both hands are needed to connect to RNA.
But the researchers found that, instead of each domain grabbing a section of the loop, only RRM1 makes contact with the RNA. RRM2 acts as support, helping organize RRM1 into the structure needed to conform to a certain section of the loop.
To confirm that the structures are key to binding, the researchers inserted mutations by changing amino acids on the surface of the domains.
Surprisingly, mutations on the far side of RRM1 — the surface not in contact with the RNA but with the RRM2 — caused decoupling at that site and substantially weakened the affinity for RNA.
Without the normal connection between the two domains, RRM1 fails to adopt the geometric shape that conforms to the RNA hairpin loop.
The researchers are further investigating how the protein transmits the effects of RRM2 to RRM1 and bind. They are also exploring the development of antagonistic agents that would disrupt the interaction of the protein with viruses.
Natural defense protein against HIV discovered

Earlier research had shown that it was possible to interfere with HIV spread but the exact molecular mechanisms had not been identified. For the first time, scientists have identified ERManI (Endoplasmic Reticulum Class I α-Mannosidase) as the essential host protein that slows the spread of HIV-1. Scientists investigated how the four ER-associated glycoside hydrolase family 47 (GH47) α-mannosidases, ERManI, and ER-degradation enhancing α-mannosidase-like (EDEM) proteins 1, 2, and 3, are involved in the HIV-1 envelope (Env) degradation process. Ectopic expression of these four α-mannosidases uncovered that only ERManI inhibited HIV-1 Env expression in a dose-dependent manner. Basically, ERManI is a host enzyme that adds sugars to proteins. The Env glycoprotein is targeted to the endoplasmic reticulum-associated protein degradation pathway for degradation after infecting cells. And ERManI was found to interact with the Env and initiate this degradation pathway.
With this discovery, ERManI has the potential as a new antiretroviral treatment option. Currently there is no cure for HIV-1 and once patients are infected, they have it for life. Current antiretroviral therapies can prolong life but cannot fully cure a patient. ERManI is different from current treatments in the sense that it can help the body protect itself.
ERManI (Endoplasmic Reticulum Class I α-Mannosidase) Is Required for HIV-1 Envelope Glycoprotein Degradation via Endoplasmic Reticulum-associated Protein Degradation Pathway (Sep 2015)
ERManI (Endoplasmic Reticulum Class I α-Mannosidase) Is Required for HIV-1 Envelope Glycoprotein Degradation via Endoplasmic Reticulum-associated Protein Degradation Pathway.
Previously, we reported that the mitochondrial translocator protein (TSPO) induces HIV-1 envelope (Env) degradation via the endoplasmic reticulum (ER)-associated protein degradation (ERAD) pathway, but the mechanism was not clear. Here we investigated how the four ER-associated glycoside hydrolase family 47 (GH47) α-mannosidases, ERManI, and ER-degradation enhancing α-mannosidase-like (EDEM) proteins 1, 2, and 3, are involved in the Env degradation process. Ectopic expression of these four α-mannosidases uncovers that only ERManI inhibits HIV-1 Env expression in a dose-dependent manner. In addition, genetic knock-out of the ERManI gene MAN1B1 using CRISPR/Cas9 technology disrupts the TSPO-mediated Env degradation. Biochemical studies show that HIV-1 Env interacts with ERManI, and between the ERManI cytoplasmic, transmembrane, lumenal stem, and lumenal catalytic domains, the catalytic domain plays a critical role in the Env-ERManI interaction. In addition, functional studies show that inactivation of the catalytic sites by site-directed mutagenesis disrupts the ERManI activity. These studies identify ERManI as a critical GH47 α-mannosidase in the ER-associated protein degradation pathway that initiates the Env degradation and suggests that its catalytic domain and enzymatic activity play an important role in this process.
T cell editing using CRISPR/Cas9 could revolutionize HIV therapeutics
September 15, 2015

Reinforcing the immune system by engineering lymphocytes to target and destroy viruses has the potential to be an effective therapy for many diseases. One potential approach to this strategy is to alter the genome of lymphocytes so that proteins that are typically hijacked by viruses are no longer present. While conceptually feasible, editing T cells has been challenging in practice; however, with the advent of mammalian cell editing using CRISPR/Cas9, T-cell editing is closer to becoming a reality.
How can CRISPR/Cas9 bring us closer to finding a cure for HIV?
In a study recently published in PNAS, scientists have optimized a protocol to introduce nucleotide replacements that would inhibit CXCR4 expression. The authors streamlined the CRISPR/Cas9 editing process by electroporating Cas9 ribonucleoproteins (RNPs) into CD4+ T cells. The RNPs, consisting of both a recombinant Cas9 enzyme and guide RNA, vastly improved editing efficiency, ultimately promoting knock-out of the CXCR4 cell-surface receptor. Taken together, these result suggest the potential of a new cell therapy approach for the fight against HIV.
Generation of knock-in primary human T cells using Cas9 ribonucleoproteins
Kathrin Schumann a , b , 1 , Steven Lin c , 1 , Eric Boyer a , b , Dimitre R. Simeonov a , b , d , Meena Subramaniam e , f , Rachel E. Gate e , f , et al. PNAS. 2015; 112(33): 10437-10442. http://dx.doi.org:/10.1073/pnas.1512503112
Significance
T-cell genome engineering holds great promise for cancer immunotherapies and cell-based therapies for HIV, primary immune deficiencies, and autoimmune diseases, but genetic manipulation of human T cells has been inefficient. We achieved efficient genome editing by delivering Cas9 protein pre-assembled with guide RNAs. These active Cas9 ribonucleoproteins (RNPs) enabled successful Cas9-mediated homology-directed repair in primary human T cells. Cas9 RNPs provide a programmable tool to replace specific nucleotide sequences in the genome of mature immune cells—a longstanding goal in the field. These studies establish Cas9 RNP technology for diverse experimental and therapeutic genome engineering applications in primary human T cells.
T-cell genome engineering holds great promise for cell-based therapies for cancer, HIV, primary immune deficiencies, and autoimmune diseases, but genetic manipulation of human T cells has been challenging. Improved tools are needed to efficiently “knock out” genes and “knock in” targeted genome modifications to modulate T-cell function and correct disease-associated mutations. CRISPR/Cas9 technology is facilitating genome engineering in many cell types, but in human T cells its efficiency has been limited and it has not yet proven useful for targeted nucleotide replacements. Here we report efficient genome engineering in human CD4+ T cells using Cas9:single-guide RNA ribonucleoproteins (Cas9 RNPs). Cas9 RNPs allowed ablation of CXCR4, a coreceptor for HIV entry. Cas9 RNP electroporation caused up to ∼40% of cells to lose high-level cell-surface expression of CXCR4, and edited cells could be enriched by sorting based on low CXCR4 expression. Importantly, Cas9 RNPs paired with homology-directed repair template oligonucleotides generated a high frequency of targeted genome modifications in primary T cells. Targeted nucleotide replacement was achieved in CXCR4 and PD-1 (PDCD1), a regulator of T-cell exhaustion that is a validated target for tumor immunotherapy. Deep sequencing of a target site confirmed that Cas9 RNPs generated knock-in genome modifications with up to ∼20% efficiency, which accounted for up to approximately one-third of total editing events. These results establish Cas9 RNP technology for diverse experimental and therapeutic genome engineering applications in primary human T cells.
Like this:
Like Loading...
Read Full Post »